Phenotypic Differentiation Within the aac(6' ) Aminoglycoside Resistance Gene Family Suggests a Novel Subtype IV of Contemporary Clinical Relevance
- PMID: 39766586
- PMCID: PMC11672645
- DOI: 10.3390/antibiotics13121196
Phenotypic Differentiation Within the aac(6' ) Aminoglycoside Resistance Gene Family Suggests a Novel Subtype IV of Contemporary Clinical Relevance
Abstract
Background: Whole genome sequencing of clinical bacterial isolates holds promise in predicting their susceptibility to antibiotic therapy, based on a detailed understanding of the phenotypic manifestation of genotypic variation. The aac(6') aminoglycoside acetyltransferase gene family is the most abundant aminoglycoside resistance determinant encountered in clinical practice. A variety of AAC(6') isozymes have been described, suggesting a phenotypic distinction between subtype I, conferring resistance to amikacin (AMK), and subtype II, conferring resistance to gentamicin (GEN) instead. However, the epidemiology and thus clinical relevance of the various and diverse isozymes and their phenotypic distinction demand systematic and contemporary re-assessment to reliably predict bacterial susceptibility to aminoglycoside antibiotics.
Methods: We analyzed the resistance gene annotations of 657,603 clinical bacterial isolates to assess the prevalence and diversity of aac(6') genes. Seventeen unique aac(6') amino acid sequences were cloned and expressed under defined promoter control in otherwise isogenic E. coli cells for phenotypic analysis with twenty distinct aminoglycoside antibiotics. A panel of clinical isolates was analyzed for the genotype-phenotype correlation of aac(6').
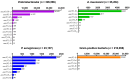
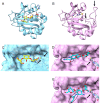
Results: An aac(6') resistance gene annotation was found in 139,236 (21.2%) of the clinical isolates analyzed. AMK resistance-conferring aac(6')-I genes dominated in Enterobacterales (28.5%). In Pseudomonas aeruginosa and Acinetobacter baumannii, a gene conferring the aac(6')-II phenotype but annotated as aac(6')-Ib4 was the most prevalent. None of the aac(6') genes were annotated as subtype III, but gene aac(6')-Ii identified in Gram-positive isolates displayed a subtype III phenotype. Genes that were annotated as aac(6')-Ib11 in Enterobacterales conferred resistance to both AMK and GEN, which we propose constitutes a novel subtype IV when applying established nomenclature. A phenotypic assessment facilitated structural re-assessment of the substrate promiscuity of AAC(6') enzymes.
Conclusions: Our study provides the most comprehensive analysis of clinically relevant aac(6') gene sequence variations to date, providing new insights into a differentiated substrate promiscuity across the genotypic spectrum of this gene family, thus translating into a critical contribution towards the development of amino acid sequence-based in silico antimicrobial susceptibility testing (AST).
Keywords: acetyltransferases; aminoglycoside antibiotics; in silico antimicrobial susceptibility testing; phenotype prediction; resistance gene annotation; surveillance; whole genome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Paul M., Carrara E., Retamar P., Tängdén T., Bitterman R., Bonomo R.A., de Waele J., Daikos G.L., Akova M., Harbarth S., et al. European Society of Clinical Microbiology and Infectious Diseases (ESCMID) guidelines for the treatment of infections caused by multidrug-resistant Gram-negative bacilli (endorsed by European society of intensive care medicine) Clin. Microbiol. Infect. 2022;28:521–547. doi: 10.1016/j.cmi.2021.11.025. - DOI - PubMed
-
- Stogios P.J., Kuhn M.L., Evdokimova E., Law M., Courvalin P., Savchenko A. Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes. ACS Infect. Dis. 2017;3:132–143. doi: 10.1021/acsinfecdis.6b00058. - DOI - PubMed
LinkOut - more resources
Full Text Sources

