Genotypic Influences on Actuators of Aerobic Performance in Tactical Athletes
- PMID: 39766802
- PMCID: PMC11675622
- DOI: 10.3390/genes15121535
Genotypic Influences on Actuators of Aerobic Performance in Tactical Athletes
Abstract
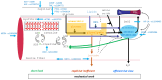
Background: This study examines genetic variations in the systemic oxygen transport cascade during exhaustive exercise in physically trained tactical athletes. Research goal: To update the information on the distribution of influence of eleven polymorphisms in ten genes, namely ACE (rs1799752), AGT (rs699), MCT1 (rs1049434), HIF1A (rs11549465), COMT (rs4680), CKM (rs8111989), TNC (rs2104772), PTK2 (rs7460 and rs7843014), ACTN3 (rs1815739), and MSTN (rs1805086)-on the connected steps of oxygen transport during aerobic muscle work.
Methods: 251 young, healthy tactical athletes (including 12 females) with a systematic physical training history underwent exercise tests, including standardized endurance running with a 12.6 kg vest. Key endurance performance metrics were assessed using ergospirometry, blood sampling, and near-infrared spectroscopy of knee and ankle extensor muscles. The influence of gene polymorphisms on the above performance metrics was analyzed using Bayesian analysis of variance.
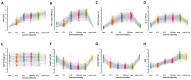
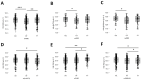
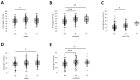
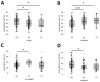
Results: Subjects exhibited good aerobic fitness (maximal oxygen uptake (VO2max): 4.3 ± 0.6 L min-1, peak aerobic power: 3.6 W ± 0.7 W kg-1). Energy supply-related gene polymorphisms rs1799752, rs4680, rs1049434, rs7843014, rs11549465, and rs8111989 did not follow the Hardy-Weinberg equilibrium. Polymorphisms in genes that regulate metabolic and contractile features were strongly associated with variability in oxygen transport and metabolism, such as body mass-related VO2 (rs7843014, rs2104772), cardiac output (rs7460), total muscle hemoglobin content (rs7460, rs4680), oxygen saturation in exercised muscle (rs1049434), and respiration exchange ratio (rs7843014, rs11549465) at first or secondary ventilatory thresholds or VO2max. Moderate influences were found for mass-related power output.
Conclusions: The posterior distribution of effects from genetic modulators of aerobic metabolism and muscle contractility mostly confirmed prior opinions in the direction of association. The observed genetic effects of rs4680 and rs1049434 indicate a crucial role of dopamine- and lactate-modulated muscle perfusion and oxygen metabolism during running, suggesting self-selection in Swiss tactical athletes.
Keywords: exercise; fatigue; gene; heart; muscle; resistance; strength; ventilation.
Conflict of interest statement
Author MF owns stocks in Company Physiogene. The funders had no role in the design of this study, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- Hornick A., Daniels C.J. Endurance Sports Medicine: A Clinical Guide. 2nd ed. Springer International Publishing; Berlin/Heidelberg, Germany: 2023. Cardiovascular Evaluation and Treatment in the Endurance Athlete; pp. 19–36.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

