Upregulation of ABLIM1 Differentiates Intrahepatic Cholangiocarcinoma from Hepatocellular Carcinoma and Both Colorectal and Pancreatic Adenocarcinoma Liver Metastases
- PMID: 39766812
- PMCID: PMC11675665
- DOI: 10.3390/genes15121545
Upregulation of ABLIM1 Differentiates Intrahepatic Cholangiocarcinoma from Hepatocellular Carcinoma and Both Colorectal and Pancreatic Adenocarcinoma Liver Metastases
Abstract
Background: Altered gene expression in cancers holds great potential to improve the diagnostics and differentiation of primary and metastatic liver cancers. In this study, the expression of the protein-coding genes ring finger protein 135 (RNF135), ephrin-B2 (EFNB2), ring finger protein 125 (RNF125), homeobox-C 4 (HOXC4), actin-binding LIM protein 1 (ABLIM1) and oncostatin M receptor (OSMR) and the long non-coding RNAs (lncRNA) prospero homeobox 1 antisense RNA 1 (PROX1-AS1) and leukemia inhibitory factor receptor antisense RNA 1 (LIFR-AS1) was investigated in hepatocellular carcinoma, cholangiocarcinoma, colorectal liver metastases and pancreatic ductal adenocarcinoma liver metastases.
Methods: This study included 149 formalin-fixed, paraffin-embedded samples from 80 patients. After RNA isolation, quantification, reverse transcription and preamplification, real-time qPCR was performed. The gene expression between different groups was calculated relative to the expression of the reference genes using the ∆∆Cq method and statistically analyzed. The expression of the genes was additionally analyzed using the AmiCA and UCSC Xena platforms.
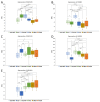
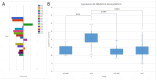
Results: In primary cancers, our results showed differential expression between primary tumors and healthy tissues for all the genes and lncRNA examined. Moreover, we found downregulation of RNF135 in hepatocellular carcinoma, downregulation of OSMR in colorectal liver metastases and upregulation of HOXC4 in cholangiocarcinoma compared to primary liver cancers and metastatic cancers. The major finding is the upregulation of ABLIM1 in cholangiocarcinoma compared to hepatocellular carcinoma, colorectal liver metastases, pancreatic ductal adenocarcinoma liver metastases and healthy liver tissue. We propose ABLIM1 as a potential biomarker that differentiates cholangiocarcinoma from other cancers and healthy liver tissue.
Conclusions: This study emphasizes the importance of understanding the differences in gene expression between healthy tissues and primary and metastatic cancers and highlights the potential use of altered gene expression as a diagnostic biomarker in these malignancies.
Keywords: ABLIM1; cholangiocarcinoma; colorectal carcinoma; colorectal liver metastases; gene expression; hepatocellular carcinoma; lncRNA; mRNA; pancreatic adenocarcinoma liver metastases; pancreatic ductal adenocarcinoma.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
DNA methylation biomarker panels for differentiating various liver adenocarcinomas, including hepatocellular carcinoma, cholangiocarcinoma, colorectal liver metastases and pancreatic adenocarcinoma liver metastases.Clin Epigenetics. 2024 Nov 4;16(1):153. doi: 10.1186/s13148-024-01766-z. Clin Epigenetics. 2024. PMID: 39497215 Free PMC article.
-
High mobility group A1 is expressed in metastatic adenocarcinoma to the liver and intrahepatic cholangiocarcinoma, but not in hepatocellular carcinoma: its potential use in the diagnosis of liver neoplasms.J Gastroenterol. 2003;38(12):1144-9. doi: 10.1007/s00535-003-1221-9. J Gastroenterol. 2003. PMID: 14714251
-
Upregulation of long noncoding RNAs LINC00941 and ABHD11-AS1 is associated with intrahepatic cholangiocarcinoma.Sci Prog. 2025 Jan-Mar;108(1):368504251330019. doi: 10.1177/00368504251330019. Epub 2025 Mar 28. Sci Prog. 2025. PMID: 40151866 Free PMC article.
-
Imaging approach to hepatocellular carcinoma, cholangiocarcinoma, and metastatic colorectal cancer.Surg Oncol Clin N Am. 2015 Jan;24(1):19-40. doi: 10.1016/j.soc.2014.09.002. Epub 2014 Oct 11. Surg Oncol Clin N Am. 2015. PMID: 25444467 Review.
-
The role of long non-coding RNA AFAP1-AS1 in human malignant tumors.Pathol Res Pract. 2018 Oct;214(10):1524-1531. doi: 10.1016/j.prp.2018.08.014. Epub 2018 Aug 20. Pathol Res Pract. 2018. PMID: 30173945 Review.
Cited by
-
Analysis of lifespan across diversity outbred mouse studies identifies multiple longevity-associated loci.Genetics. 2025 Aug 6;230(4):iyaf081. doi: 10.1093/genetics/iyaf081. Genetics. 2025. PMID: 40326784 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

