Exploring Immune Cell Infiltration and Small Molecule Compounds for Ulcerative Colitis Treatment
- PMID: 39766817
- PMCID: PMC11728156
- DOI: 10.3390/genes15121548
Exploring Immune Cell Infiltration and Small Molecule Compounds for Ulcerative Colitis Treatment
Abstract
Background/objectives: Ulcerative colitis (UC) is a chronic inflammatory bowel disease (IBD) with a relapsing nature and complex etiology. Bioinformatics analysis has been widely applied to investigate various diseases. This study aimed to identify crucial differentially expressed genes (DEGs) and explore potential therapeutic agents for UC.
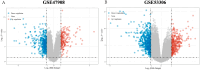
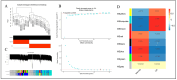
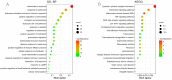
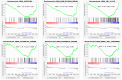
Methods: The GSE47908 and GSE55306 colon tissue transcriptome gene datasets were downloaded from the Gene Expression Omnibus-NCBI (GEO) database. GEO2R and Gene Set Enrichment Analysis (GSEA) were used to screen for DEGs in patients with UC compared to the normal population based on weighted gene co-expression network analysis (WGCNA). GO-BP analysis and KEGG enrichment analysis were performed on the intersecting differential genes via the Metascape website, while hub genes were analyzed by STRING11.0 and Cytoscape3.7.1. The expression of hub genes was verified in the dataset GSE38713 colon tissue specimens. Finally, the gene expression profiles of the validation set were analyzed by immuno-infiltration through the ImmuCellAI online tool, and the CMap database was used to screen for negatively correlated small molecule compounds.
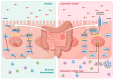
Results: A total of 595 and 926 genes were screened by analysis of GSE47908 and GSE55306 datasets, respectively. Combined WGCNA hub module intersection yielded 12 hub genes (CXCL8, IL1β, CXCL1, CCL20, CXCL2, CXCR2, LCN2, SELL, AGT, LILRB3, MMP3, IDO1) associated with the pathogenesis of UC. GSEA analysis yielded intersecting pathways for both datasets (colorectal cancer pathway, base excision repair, cell cycle, apoptosis). GO-BP and KEGG enrichment analyses were performed to obtain key biological processes (inflammatory response, response to bacteria, leukocyte activation involved in the immune response, leukocyte-cell adhesion, apoptosis, positive regulation of immune effector processes) and key signaling pathways (cytokine-cytokine receptor interactions, IBD, NOD-like receptor signaling pathways). The immune cell infiltration analysis suggested that the incidence of UC was mainly related to the increase in CD4+T cells, depletion of T cells, T follicular helper cells, natural killer cells, γδ T cells and the decrease in CD8 naive T cells, helper T cells 17 and effector T cells. The CMap database results showed that small molecule compounds such as vorinostat, roxarsone, and wortmannin may be therapeutic candidates for UC.
Conclusions: This study not only aids in early prediction and prevention but also provides novel insights into the pathogenesis and treatment of UC.
Keywords: bioinformatics; differentially expressed genes; gene chip; potential therapeutic drugs; ulcerative colitis.
Conflict of interest statement
Author Yi Lu was employed by the company Shanghai Tufeng Pharmaceutical Technology Co. Graphical Abstract and Figure 9 were created by Figdraw. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Identification of shared gene signatures and molecular mechanisms between chronic kidney disease and ulcerative colitis.Front Immunol. 2023 Feb 13;14:1078310. doi: 10.3389/fimmu.2023.1078310. eCollection 2023. Front Immunol. 2023. PMID: 36860851 Free PMC article.
-
Exploring Pyroptosis-related Signature Genes and Potential Drugs in Ulcerative Colitis by Transcriptome Data and Animal Experimental Validation.Inflammation. 2024 Dec;47(6):2057-2076. doi: 10.1007/s10753-024-02025-2. Epub 2024 Apr 24. Inflammation. 2024. PMID: 38656456
-
Identification of key genes and biological processes contributing to colitis associated dysplasia in ulcerative colitis.PeerJ. 2021 Apr 27;9:e11321. doi: 10.7717/peerj.11321. eCollection 2021. PeerJ. 2021. PMID: 33987007 Free PMC article.
-
Identification of Immune-Related Gene Signature and Prediction of CeRNA Network in Active Ulcerative Colitis.Front Immunol. 2022 Mar 22;13:855645. doi: 10.3389/fimmu.2022.855645. eCollection 2022. Front Immunol. 2022. PMID: 35392084 Free PMC article.
-
Unveiling the key genes, environmental toxins, and drug exposures in modulating the severity of ulcerative colitis: a comprehensive analysis.Front Immunol. 2023 Jul 19;14:1162458. doi: 10.3389/fimmu.2023.1162458. eCollection 2023. Front Immunol. 2023. PMID: 37539055 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- CSC202308310088/China Scholarship Council
- CSC202108440109/China Scholarship Council
- YC-2023-0408/Demonstration Pilot Project of Traditional Chinese Medicine Inheritance and Innovation in Pudong New District shanghai-Flagship Community Health Service Centre for Integrative Chinese and Western Medicine
- Pudong New District Health Committee/2024 High-level Talent Mentor Teaching Young Talents Project of Shanghai
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

