From Omics to Multi-Omics: A Review of Advantages and Tradeoffs
- PMID: 39766818
- PMCID: PMC11675490
- DOI: 10.3390/genes15121551
From Omics to Multi-Omics: A Review of Advantages and Tradeoffs
Abstract
Bioinformatics is a rapidly evolving field charged with cataloging, disseminating, and analyzing biological data. Bioinformatics started with genomics, but while genomics focuses more narrowly on the genes comprising a genome, bioinformatics now encompasses a much broader range of omics technologies. Overcoming barriers of scale and effort that plagued earlier sequencing methods, bioinformatics adopted an ambitious strategy involving high-throughput and highly automated assays. However, as the list of omics technologies continues to grow, the field of bioinformatics has changed in two fundamental ways. Despite enormous success in expanding our understanding of the biological world, the failure of bulk methods to account for biologically important variability among cells of the same or different type has led to a major shift toward single-cell and spatially resolved omics methods, which attempt to disentangle the conflicting signals contained in heterogeneous samples by examining individual cells or cell clusters. The second major shift has been the attempt to integrate two or more different classes of omics data in a single multimodal analysis to identify patterns that bridge biological layers. For example, unraveling the cause of disease may reveal a metabolite deficiency caused by the failure of an enzyme to be phosphorylated because a gene is not expressed due to aberrant methylation as a result of a rare germline variant. Conclusions: There is a fine line between superficial understanding and analysis paralysis, but like a detective novel, multi-omics increasingly provides the clues we need, if only we are able to see them.
Keywords: bioinformatics; epigenomics; glycoinformatics; lipidomics; metabolomics; proteomics; single-cell RNA sequencing; spatially resolved transcriptomics; transcriptomics.
Conflict of interest statement
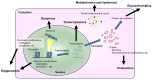
The authors declare no conflicts of interest. Figure 2 was partially created using
Figures

References
-
- Rosenberg E. It’s in Your DNA: From Discovery to Structure, Function and Role in Evolution, Cancer and Aging. Academic Press; London, UK: 2017. 199p pp. xvii.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

