Exploration of Key Regulatory Factors in Mesenchymal Stem Cell Continuous Osteogenic Differentiation via Transcriptomic Analysis
- PMID: 39766835
- PMCID: PMC11675713
- DOI: 10.3390/genes15121568
Exploration of Key Regulatory Factors in Mesenchymal Stem Cell Continuous Osteogenic Differentiation via Transcriptomic Analysis
Abstract
Background/objectives: Mesenchymal stem cells (MSCs) possess the remarkable ability to differentiate into various cell types, including osteoblasts. Understanding the molecular mechanisms governing MSC osteogenic differentiation is crucial for advancing clinical applications and our comprehension of complex disease processes. However, the key biological molecules regulating this process remain incompletely understood.
Methods: In this study, we conducted systematic re-analyses of published high-throughput transcriptomic datasets to identify and validate key biological molecules that dynamically regulate MSC osteogenic differentiation. Our approach involved a comprehensive analysis of gene expression patterns across human tissues, followed by the rigorous experimental validation of the identified candidates.
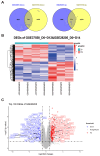
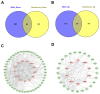
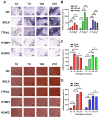
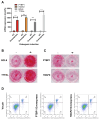
Results: Through integrated analytical and experimental approaches, we utilized high-throughput transcriptomics to identify four critical regulators of MSC osteogenic differentiation: PTBP1, H2AFZ, BCL6, and TTPAL (C20ORF121). Among these, PTBP1 and H2AFZ functioned as positive regulators, while BCL6 and TTPAL acted as negative regulators in osteogenesis. The regulatory roles of these genes in osteogenesis were further validated via overexpression experiments.
Conclusions: Our findings advance our understanding of MSC differentiation fate determination and open new therapeutic possibilities for bone-related disorders. The identification of these regulators provides a foundation for developing targeted interventions in regenerative medicine.
Keywords: MSCs; dynamic regulation; lineage change; osteogenesis; transcriptomic sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Zhang H., Xu R., Li B., Xin Z., Ling Z., Zhu W., Li X., Zhang P., Fu Y., Chen J., et al. LncRNA NEAT1 controls the lineage fates of BMSCs during skeletal aging by impairing mitochondrial function and pluripotency maintenance. Cell Death Differ. 2022;29:351–365. doi: 10.1038/s41418-021-00858-0. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

