Genetic Analysis of the Peach SnRK1β3 Subunit and Its Function in Transgenic Tomato Plants
- PMID: 39766841
- PMCID: PMC11675834
- DOI: 10.3390/genes15121574
Genetic Analysis of the Peach SnRK1β3 Subunit and Its Function in Transgenic Tomato Plants
Abstract
Background/objectives: The sucrose non-fermentation-related kinase 1 (SnRK1) protein complex in plants plays an important role in energy metabolism, anabolism, growth, and stress resistance. SnRK1 is a heterotrimeric complex. The SnRK1 complex is mainly composed of α, β, βγ, and γ subunits. Studies on plant SnRK1 have primarily focused on the functional α subunit, with the β regulatory subunit remaining relatively unexplored. The present study aimed to elucidate the evolutionary relationship, structural prediction, and interaction with the core α subunit of peach SnRK1β3 (PpSnRK1) subunit.
Methods: Bioinformatics analysis of PpSnRK1 was performed through software and website. We produced transgenic tomato plants overexpressing PpSnRK1 (OEPpSnRK1). Transcriptome analysis was performed on OEPpSnRK1 tomatoes. We mainly tested the growth index and drought resistance of transgenic tomato plants.
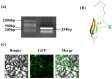
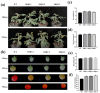
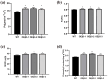
Results: The results showed that PpSnRK1 has a 354 bp encoded protein sequence (cds), which is mainly located in the nucleus and cell membrane. Phylogenetic tree analysis showed that PpSnRK1β3 has similar domains to other woody plants. Transcriptome analysis of OEPpSnRK1β3 showed that PpSnRK1β3 is widely involved in biosynthetic and metabolic processes. Functional analyses of these transgenic plants revealed prolonged growth periods, enhanced growth potential, improved photosynthetic activity, and superior drought stress tolerance.
Conclusions: The study findings provide insight into the function of the PpSnRK1 subunit and its potential role in regulating plant growth and drought responses. This comprehensive analysis of PpSnRK1 will contribute to further enhancing our understanding of the plant SnRK1 protein complex.
Keywords: PpSnRK1β3; peach; transgenic tomato plants.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Alderson A., Sabelli P.A., Dickinson J.R., Cole D., Richardson M., Kreis M., Shewry P.R., Halford N.G. Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc. Natl. Acad. Sci. USA. 1991;88:8602–8605. doi: 10.1073/pnas.88.19.8602. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

