HROB Is Implicated in DNA Replication
- PMID: 39766854
- PMCID: PMC11675949
- DOI: 10.3390/genes15121587
HROB Is Implicated in DNA Replication
Abstract
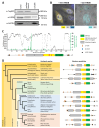
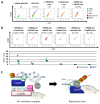
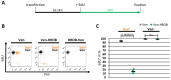
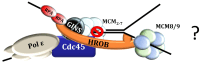
DNA replication represents a series of precisely regulated events performed by a complex protein machinery that guarantees accurate duplication of the genetic information. Since DNA replication is permanently faced by a variety of exogenous and endogenous stressors, DNA damage response, repair and replication must be closely coordinated to maintain genomic integrity. HROB has been identified recently as a binding partner and activator of the Mcm8/9 helicase involved in DNA interstrand crosslink (ICL) repair. We identified HROB independently as a nuclear protein whose expression is co-regulated with various DNA replication factors. Accordingly, the HROB protein level showed a maximum in S phase and a downregulation in quiescence. Structural prediction and homology searches revealed that HROB is a largely intrinsically disordered protein bearing a helix-rich region and a canonical oligonucleotide/oligosaccharide-binding-fold motif that originated early in eukaryotic evolution. Employing a flow cytometry Förster resonance energy transfer (FRET) assay, we detected associations between HROB and proteins of the DNA replication machinery. Moreover, ectopic expression of HROB protein led to an almost complete shutdown of DNA replication. The available data imply a function for HROB during DNA replication across barriers such as ICLs.
Keywords: DNA interstrand crosslink (ICL) repair; DNA replication; Förster resonance energy transfer (FRET); HROB; protein overexpression; soluble protein fragment identification.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Control of homologous recombination by the HROB-MCM8-MCM9 pathway.Genes Dev. 2019 Oct 1;33(19-20):1397-1415. doi: 10.1101/gad.329508.119. Epub 2019 Aug 29. Genes Dev. 2019. PMID: 31467087 Free PMC article.
-
Mechanism of DNA unwinding by MCM8-9 in complex with HROB.Nat Commun. 2024 Apr 27;15(1):3584. doi: 10.1038/s41467-024-47936-8. Nat Commun. 2024. PMID: 38678026 Free PMC article.
-
C17orf53 is identified as a novel gene involved in inter-strand crosslink repair.DNA Repair (Amst). 2020 Nov;95:102946. doi: 10.1016/j.dnarep.2020.102946. Epub 2020 Aug 15. DNA Repair (Amst). 2020. PMID: 32853826 Free PMC article.
-
Orchestrating the nucleases involved in DNA interstrand cross-link (ICL) repair.Cell Cycle. 2011 Dec 1;10(23):3999-4008. doi: 10.4161/cc.10.23.18385. Epub 2011 Dec 1. Cell Cycle. 2011. PMID: 22101340 Free PMC article. Review.
-
Fanconi anemia-independent DNA inter-strand crosslink repair in eukaryotes.Prog Biophys Mol Biol. 2020 Dec;158:33-46. doi: 10.1016/j.pbiomolbio.2020.08.005. Epub 2020 Aug 30. Prog Biophys Mol Biol. 2020. PMID: 32877700 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

