Mechanisms, Machinery, and Dynamics of Chromosome Segregation in Zea mays
- PMID: 39766873
- PMCID: PMC11675298
- DOI: 10.3390/genes15121606
Mechanisms, Machinery, and Dynamics of Chromosome Segregation in Zea mays
Abstract
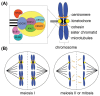
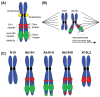
Zea mays (maize) is both an agronomically important crop and a powerful genetic model system with an extensive molecular toolkit and genomic resources. With these tools, maize is an optimal system for cytogenetic study, particularly in the investigation of chromosome segregation. Here, we review the advances made in maize chromosome segregation, specifically in the regulation and dynamic assembly of the mitotic and meiotic spindle, the inheritance and mechanisms of the abnormal chromosome variant Ab10, the regulation of chromosome-spindle interactions via the spindle assembly checkpoint, and the function of kinetochore proteins that bridge chromosomes and spindles. In this review, we discuss these processes in a species-specific context including features that are both conserved and unique to Z. mays. Additionally, we highlight new protein structure prediction tools and make use of these tools to identify several novel kinetochore and spindle assembly checkpoint proteins in Z. mays.
Keywords: Zea mays; chromosome; kinetochore; meiosis; meiotic drive; mitosis; spindle; spindle assembly checkpoint.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- McClintock B. Cytological observations of deficiencies involving known genes, translocations and an inversion in Zea mays. Mo. Agric. Exp. Stn. Res. Bull. 1931;163:1–30.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

