Genomic Analysis of Talaromyces verruculosus SJ9: An Efficient Tetracycline-, Enrofloxacin-, and Tylosin-Degrading Fungus
- PMID: 39766911
- PMCID: PMC11675779
- DOI: 10.3390/genes15121643
Genomic Analysis of Talaromyces verruculosus SJ9: An Efficient Tetracycline-, Enrofloxacin-, and Tylosin-Degrading Fungus
Abstract
Background/objectives: Many fungi related to Talaromyces verruculosus can degrade a wide range of pollutants and are widely distributed globally. T. verruculosus SJ9 was enriched from fresh strawberry inter-root soil to yield fungi capable of degrading tetracycline, enrofloxacin, and tylosin.
Methods: T. verruculosus SJ9 genome was sequenced, assembled, and annotated in this study utilizing bioinformatics software, PacBio, and the Illumina NovaSeq PE150 technology.
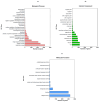
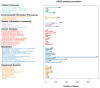
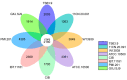
Results: The genome size is 40.6 Mb, the N50 scaffold size is 4,534,389 bp, and the predicted number of coding genes is 8171. The T. verruculosus TS63-9 genome has the highest resemblance to the T. verruculosus SJ9 genome, according to a comparative genomic analysis of seven species. In addition, we annotated many genes encoding antibiotic-degrading enzymes in T. verruculosus SJ9 through genomic databases, which also provided strong evidence for its ability to degrade antibiotics.
Conclusions: Through the correlation analysis of the whole-genome data of T. verruculosus SJ9, we identified a number of genes capable of encoding antibiotic-degrading enzymes in its gene function annotation database. These antibiotic-related enzymes provide some evidence that T. verruculosus SJ9 can degrade fluoroquinolone antibiotics, tetracycline antibiotics, and macrolide antibiotics. In summary, the complete genome sequence of T. verruculosus SJ9 has now been published, and this resource constitutes a significant dataset that will inform forthcoming transcriptomic, proteomic, and metabolic investigations of this fungal species. In addition, genomic studies of other filamentous fungi can utilize it as a reference. Thanks to the discoveries made in this study, the future application of this fungus in industrial production will be more rapid.
Keywords: T. verruculosus; antibiotic degradation; functional annotation; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Draft genome sequence of Talaromyces verruculosus ("Penicillium verruculosum") strain TS63-9, a fungus with great potential for industrial production of polysaccharide-degrading enzymes.J Biotechnol. 2016 Feb 10;219:5-6. doi: 10.1016/j.jbiotec.2015.12.017. Epub 2015 Dec 18. J Biotechnol. 2016. PMID: 26707807
-
Genome sequencing and analysis of Talaromyces pinophilus provide insights into biotechnological applications.Sci Rep. 2017 Mar 28;7(1):490. doi: 10.1038/s41598-017-00567-0. Sci Rep. 2017. PMID: 28352091 Free PMC article.
-
Unveiling the optimal parameters for cellulolytic characteristics of Talaromyces verruculosus SGMNPf3 and its secretory enzymes.J Appl Microbiol. 2015 Jul;119(1):88-98. doi: 10.1111/jam.12816. Epub 2015 May 18. J Appl Microbiol. 2015. PMID: 25833715
-
Efficient degradation of tylosin by Kurthia gibsonii TYL-A1: performance, pathway, and genomics study.Microbiol Spectr. 2025 Jun 3;13(6):e0002525. doi: 10.1128/spectrum.00025-25. Epub 2025 Apr 29. Microbiol Spectr. 2025. PMID: 40298383 Free PMC article.
-
Talaromyces marneffei Genomic, Transcriptomic, Proteomic and Metabolomic Studies Reveal Mechanisms for Environmental Adaptations and Virulence.Toxins (Basel). 2017 Jun 13;9(6):192. doi: 10.3390/toxins9060192. Toxins (Basel). 2017. PMID: 28608842 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

