House Mice in the Atlantic Region: Genetic Signals of Their Human Transport
- PMID: 39766912
- PMCID: PMC11675120
- DOI: 10.3390/genes15121645
House Mice in the Atlantic Region: Genetic Signals of Their Human Transport
Abstract
Background/objectives: The colonization history of house mice reflects the maritime history of humans that passively transported them worldwide. We investigated western house mouse colonization in the Atlantic region through studies of mitochondrial D-loop DNA sequences from modern specimens.
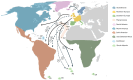
Methods: We assembled a dataset of 758 haplotypes derived from 2765 mice from 47 countries/oceanic archipelagos (a combination of new and published data). Our maximum likelihood phylogeny recovered five previously identified clades, and we used the haplotype affinities within the phylogeny to infer house mouse colonization history, employing statistical tests and indices. From human history, we predefined four European source areas for mice in the Atlantic region (Northern Europe excluding Scandinavia, Southern Europe, Scandinavia, and Macaronesia) and we investigated the colonization from these source areas to different geographic areas in the Atlantic region.
Results: Our inferences suggest mouse colonization of Scandinavia itself from Northern Europe, and Macaronesia from both Southern Europe and Scandinavia/Germany (the latter likely representing the transport of mice by Vikings). Mice on North Atlantic islands apparently derive primarily from Scandinavia, while for South Atlantic islands, North America, and Sub-Saharan Africa, the clearest source is Northern Europe, although mice on South Atlantic islands also had genetic inputs from Macaronesia and Southern Europe (for Tristan da Cunha). Macaronesia was a stopover for Atlantic voyages, creating an opportunity for mouse infestation. Mice in Latin America also apparently had multiple colonization sources, with a strong Southern European signal but also input from Northern Europe and/or Macaronesia.
Conclusions: D-loop sequences help discern the broad-scale colonization history of house mice and new perspectives on human history.
Keywords: Age of Discovery; D-loop; Mus musculus domesticus; Vikings; colonization history; phylogeography.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Hulme-Beaman A., Dobney K., Cucchi T., Searle J.B. An ecological and evolutionary framework for commensalism in anthropogenic environments. Trends Ecol. Evol. 2016;31:633–645. - PubMed
-
- Kristensen B.R. Rethinking domestication pathways in the context of anthrodependency. In: Braverman I., editor. More-than-One Health: Human, Animals and the Environment Post-COVID. Routledge; London, UK: 2022. pp. 193–208.
-
- Munshi-South J., Garcia J.A., Orton D., Phifer-Rixey M. The evolutionary history of wild and domestic brown rats (Rattus norvegicus) Science. 2024;385:1292–1297. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

