Exploring the Role of FICD, a New Potential Gene Involved in Borderline Intellectual Functioning, Psychological and Metabolic Disorders
- PMID: 39766922
- PMCID: PMC11727805
- DOI: 10.3390/genes15121655
Exploring the Role of FICD, a New Potential Gene Involved in Borderline Intellectual Functioning, Psychological and Metabolic Disorders
Abstract
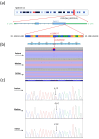
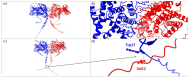
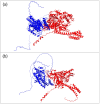
Background/Objectives: AMPylation is a post-translational modification involving the transfer of adenosine monophosphate (AMP) from adenosine triphosphate (ATP) to target proteins, serving as a critical regulatory mechanism in cellular functions. This study aimed to expand the phenotypic spectrum associated with mutations in the FICD gene, which encodes an adenyltransferase enzyme involved in both AMPylation and deAMPylation. Methods: A clinical evaluation was conducted on a patient presenting with a complex clinical profile. Whole-exome sequencing (WES) was performed to identify potential genetic variants contributing to the observed phenotype. Results: The patient exhibited borderline intellectual functioning (BIF), acanthosis, abdominal muscle hypotonia, anxiety, depression, obesity, and optic nerve subatrophy. WES revealed a de novo missense variant, c.1295C>T p.Ala432Val, in the FICD gene. This variant, classified as of uncertain significance, is located in the highly conserved region TLLFATTEY (aa 428-436), suggesting a potential impact on protein function. Conclusions: These findings highlight the importance of the FICD gene in diverse clinical manifestations and emphasize the need for further studies to elucidate the genetic mechanisms underlying these phenotypes. Continued research is essential to improve our understanding of FICD-related conditions.
Keywords: AMPylation; adenyltransferase; deAMPylation; next-generation sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Catalytic deAMPylation in AMPylation-inhibitory/assistant forms of FICD protein.Front Chem. 2023 Jan 25;11:1077188. doi: 10.3389/fchem.2023.1077188. eCollection 2023. Front Chem. 2023. PMID: 36762200 Free PMC article.
-
BiP inactivation due to loss of the deAMPylation function of FICD causes a motor neuron disease.Genet Med. 2022 Dec;24(12):2487-2500. doi: 10.1016/j.gim.2022.08.019. Epub 2022 Sep 22. Genet Med. 2022. PMID: 36136088
-
Structures of a deAMPylation complex rationalise the switch between antagonistic catalytic activities of FICD.Nat Commun. 2021 Aug 18;12(1):5004. doi: 10.1038/s41467-021-25076-7. Nat Commun. 2021. PMID: 34408154 Free PMC article.
-
Clinical Features and Genetic Characteristics of XLID Patients With KDM5C Gene Mutations: Insights on Phenotype-Genotype Correlations From 175 Previous Cases and Identification of a Novel Variant.Mol Genet Genomic Med. 2025 Jan;13(1):e70057. doi: 10.1002/mgg3.70057. Mol Genet Genomic Med. 2025. PMID: 39835750 Free PMC article. Review.
-
Exome sequencing identifies a de novo SCN2A mutation in a patient with intractable seizures, severe intellectual disability, optic atrophy, muscular hypotonia, and brain abnormalities.Epilepsia. 2014 Apr;55(4):e25-9. doi: 10.1111/epi.12554. Epub 2014 Mar 1. Epilepsia. 2014. PMID: 24579881 Review.
Cited by
-
Isolation and Identification of Inter-Correlated Genes from the Invasive Sun Corals Tubastraea Coccinea and Tubastraea Tagusensis (Scleractinia, Cnidaria).Int J Mol Sci. 2025 Jul 26;26(15):7235. doi: 10.3390/ijms26157235. Int J Mol Sci. 2025. PMID: 40806368 Free PMC article.
-
Potential Role of ABCF2 Gene in Pudendal Nerve Neuropathy and Interstitial Cystitis.Genes (Basel). 2025 Feb 26;16(3):281. doi: 10.3390/genes16030281. Genes (Basel). 2025. PMID: 40149433 Free PMC article.
References
-
- Utsumi R., Kusafuka S., Nakayama T., Tanaka K., Takayanagi Y., Takahashi H., Noda M., Kawamukai M. Stationary Phase-Specific Expression of the Fic Gene in Escherichia coli K-12 Is Controlled by the RpoS Gene Product (σ38) FEMS Microbiol. Lett. 1993;113:273–278. doi: 10.1016/0378-1097(93)90217-P. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

