Integrated Transcriptome and Metabolome Analysis Reveals Mechanism of Flavonoid Synthesis During Low-Temperature Storage of Sweet Corn Kernels
- PMID: 39766968
- PMCID: PMC11727310
- DOI: 10.3390/foods13244025
Integrated Transcriptome and Metabolome Analysis Reveals Mechanism of Flavonoid Synthesis During Low-Temperature Storage of Sweet Corn Kernels
Abstract
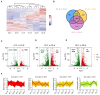
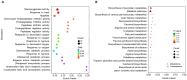
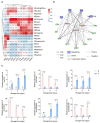
Sweet corn is a globally important food source and vegetable renowned for its rich nutritional content. However, post-harvest quality deterioration remains a significant challenge due to sweet corn's high sensitivity to environmental factors. Currently, low-temperature storage is the primary method for preserving sweet corn; however, the molecular mechanisms involved in this process remain unclear. In this study, kernels stored at different temperatures (28 °C and 4 °C) for 1, 3, and 5 days after harvest were collected for physiological and transcriptomic analysis. Low temperature storage significantly improved the PPO and SOD activity in sweet corn kernels compared to storage at a normal temperature. A total of 1993 common differentially expressed genes (DEGs) were identified in kernels stored at low temperatures across all three time points. Integrated analysis of transcriptomic and previous metabolomic data revealed that low temperature storage significantly affected flavonoid biosynthesis. Furthermore, 11 genes involved in flavonoid biosynthesis exhibited differential expression across the three storage periods, including CHI, HCT, ANS, F3'H, F3'5'H, FLS, and NOMT, with Eriodictyol, Myricetin, and Hesperetin-7-O-glucoside among the key flavonoids. Correlation analysis revealed three AP2/ERF-ERF transcription factors (EREB14, EREB182, and EREB200) as potential regulators of flavonoid biosynthesis during low temperature treatment. These results enhance our understanding of the mechanisms of flavonoid synthesis in sweet corn kernels during low-temperature storage.
Keywords: flavonoid synthesis; integrated analysis; low-temperature storage; sweet corn; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Hu Y., Colantonio V., Muller B.S.F., Leach K.A., Nanni A., Finegan C., Wang B., Baseggio M., Newton C.J., Juhl E.M., et al. Genome assembly and population genomic analysis provide insights into the evolution of modern sweet corn. Nat. Commun. 2021;12:1227. doi: 10.1038/s41467-021-21380-4. - DOI - PMC - PubMed
-
- Jan B., Anwar Bhat M., Bhat T.A., Yaqoob M., Nazir A., Ashraf Bhat M., Mir A.H., Wani F.J., Kumar Singh J., Kumar R., et al. Evaluation of seedling age and nutrient sources on phenology, yield and agrometeorological indices for sweet corn (Zea mays saccharata L.) Saudi J. Biol. Sci. 2022;29:735–742. doi: 10.1016/j.sjbs.2021.10.010. - DOI - PMC - PubMed
Grants and funding
- 2023YFD1201105/National Key Research and Development Program
- 23ZYCGSN00880/Tianjin Science and Technology Assistance project
- KLIBMC2304/Tianjin Key Laboratory of Intelligent Breeding of Major Crops
- CARS-02-85/China Agriculture Research System
- No. 102/provincial rural revitalization strategy special project of Guangdong in 2024
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

