Research on Lipidomic Profiling and Biomarker Identification for Osteonecrosis of the Femoral Head
- PMID: 39767733
- PMCID: PMC11673004
- DOI: 10.3390/biomedicines12122827
Research on Lipidomic Profiling and Biomarker Identification for Osteonecrosis of the Femoral Head
Abstract
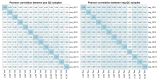
Objectives: Abnormal lipid metabolism is increasingly recognized as a contributing factor to the development of osteonecrosis of the femoral head (ONFH). This study aimed to explore the lipidomic profiles of ONFH patients, focusing on distinguishing between traumatic ONFH (TONFH) and non-traumatic ONFH (NONFH) subtypes and identifying potential biomarkers for diagnosis and understanding pathogenesis. Methods: Plasma samples were collected from 92 ONFH patients (divided into TONFH and NONFH subtypes) and 33 healthy normal control (NC) participants. Lipidomic profiling was performed using ultra-high performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS). Data analysis incorporated a machine learning-based feature selection method, least absolute shrinkage and selection operator (LASSO) regression, to identify significant lipid biomarkers. Results: Distinct lipidomic signatures were observed in both TONFH and NONFH groups compared to the NC group. LASSO regression identified 11 common lipid biomarkers that signify shared metabolic disruptions in both ONFH subtypes, several of which exhibited strong diagnostic performance with areas under the curve (AUCs) > 0.7. Additionally, subtype-specific lipid markers unique to TONFH and NONFH were identified, providing insights into the differential pathophysiological mechanisms underlying these subtypes. Conclusions: This study highlights the importance of lipidomic profiling in understanding ONFH-associated metabolic disorders and demonstrates the utility of machine learning approaches, such as LASSO regression, in high-dimensional data analysis. These findings not only improve disease characterization but also facilitate the discovery of diagnostic and mechanistic biomarkers, paving the way for more personalized therapeutic strategies in ONFH.
Keywords: LASSO; diagnostic biomarker; feature selection; lipidomic profile; osteonecrosis of the femoral head.
Conflict of interest statement
The authors declare that they have no conflicts of interests.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources

