Genomic Medicine in the Developing World: Cancer Spectrum, Cumulative Risk and Survival Outcomes for Lynch Syndrome Variant Heterozygotes with Germline Pathogenic Variants in the MLH1 and MSH2 Genes
- PMID: 39767815
- PMCID: PMC11672899
- DOI: 10.3390/biomedicines12122906
Genomic Medicine in the Developing World: Cancer Spectrum, Cumulative Risk and Survival Outcomes for Lynch Syndrome Variant Heterozygotes with Germline Pathogenic Variants in the MLH1 and MSH2 Genes
Abstract
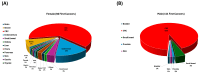
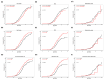
Background: Although genetic testing has improved our ability to diagnose Lynch syndrome (LS), there is still limited information on the extent of variations in the clinical and genetic landscape among LS variant heterozygotes (LSVH) in Africa. We sought to investigate the cancer spectrum, cumulative risk, and survival outcomes of LSVH with pathogenic/likely pathogenic variants (P/LPVs) in the MLH1 and MSH2 genes using a LS registry in South Africa over the last 30 years. Methods: A retrospective study was conducted to retrieve demographic, clinical, and genetic data of all LSVH with P/LPVs in the MLH1 and MSH2 genes from our LS registry. Genetic data were analyzed according to cancer spectrum, cumulative risk, and crude survival. We used the Chi-squared and t-test to assess differences between groups, and Kaplan-Meier survival analyses were used to analyze the cumulative risk and crude survival outcomes. A p-value < 0.05 at a 95% confidence interval was considered statistically significant. Results: We analyzed a total of 577 LSVH from 109 families. About 450 (78%) and 127 (22%) LSVH harbored a disease-causing mutation in MLH1 and MSH2, respectively. A South African founder PV (MLH1:c.1528C>T) accounted for 74% (n = 426) of all LSVH. CRC was the most common diagnosed cancer in both MLH1 and MSH2 LSVH. MLH1 LSVH had a younger age at cancer diagnosis than MSH2 LSVH (43 vs. 47 years, respectively, p = 0.015). Extracolonic cancers were predominantly higher in female LSVH (n = 33, 35%) than in male LSVH (n = 8, 7%) with the MLH1:c.1528C>T founder PV. The cumulative risk of any cancer and CRC at any age was higher in MLH1 LSVH than in MSH2 LSVH (p = 0.020 and p = 0.036, respectively). LSVH with the MLH1:c.1528C>T PV had a better 10-year overall survival after the first cancer diagnosis, particularly for CRC. Conclusions: LSVH with P/LPVs in the MLH1 and MSH2 genes exhibited significant gene- and sex-specific differences in cancer spectrum, cumulative risk and survival outcomes. Cancer risk and survival estimates described in this study can be used to guide surveillance and genetic counselling for LSVH in our population.
Keywords: Lynch syndrome registry; South African founder variant; colorectal cancer; crude survival; cumulative risk; extracolonic cancer; genetic variants; pathogenic variants.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Influence of Genetic Polymorphisms on the Age at Cancer Diagnosis in a Homogenous Lynch Syndrome Cohort of Individuals Carrying the MLH1:c.1528C>T South African Founder Variant.Biomedicines. 2024 Sep 27;12(10):2201. doi: 10.3390/biomedicines12102201. Biomedicines. 2024. PMID: 39457514 Free PMC article.
-
Human Leukocyte Antigen-Allelic Variations May Influence the Age at Cancer Diagnosis in Lynch Syndrome.J Pers Med. 2024 May 27;14(6):575. doi: 10.3390/jpm14060575. J Pers Med. 2024. PMID: 38929796 Free PMC article.
-
Germline variants screening of MLH1, MSH2, MSH6 and PMS2 genes in 64 Algerian Lynch syndrome families: The first nationwide study.Ann Hum Genet. 2022 Nov;86(6):328-352. doi: 10.1111/ahg.12482. Epub 2022 Sep 8. Ann Hum Genet. 2022. PMID: 36073783
-
The Clinical Outcomes Among Patients Under 60 Years Old with Lynch Syndrome: Variations Based on Different Mutation Patterns.Int J Mol Sci. 2025 Apr 4;26(7):3383. doi: 10.3390/ijms26073383. Int J Mol Sci. 2025. PMID: 40244260 Free PMC article. Review.
-
Lynch Syndrome-Impact of the Type of Deficient Mismatch Repair Gene Mutation on Diagnosis, Clinical Presentation, Surveillance and Therapeutic Approaches.Medicina (Kaunas). 2025 Jan 14;61(1):120. doi: 10.3390/medicina61010120. Medicina (Kaunas). 2025. PMID: 39859102 Free PMC article. Review.
References
-
- Tsaousis G.N., Papadopoulou E., Apessos A., Agiannitopoulos K., Pepe G., Kampouri S., Diamantopoulos N., Floros T., Iosifidou R., Katopodi O., et al. Analysis of hereditary cancer syndromes by using a panel of genes: Novel and multiple pathogenic mutations. BMC Cancer. 2019;19:535. doi: 10.1186/s12885-019-5756-4. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

