Second-Generation Antipsychotics Induce Metabolic Disruption in Adipose Tissue-Derived Mesenchymal Stem Cells Through an aPKC-Dependent Pathway
- PMID: 39768174
- PMCID: PMC11674800
- DOI: 10.3390/cells13242084
Second-Generation Antipsychotics Induce Metabolic Disruption in Adipose Tissue-Derived Mesenchymal Stem Cells Through an aPKC-Dependent Pathway
Abstract
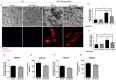
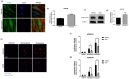
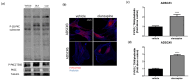
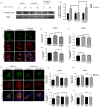
Metabolic syndrome (MetS) is a cluster of metabolic abnormalities, including visceral obesity, dyslipidemia, and insulin resistance. In this regard, visceral white adipose tissue (vWAT) plays a critical role, influencing energy metabolism, immunomodulation, and oxidative stress. Adipose-derived stem cells (ADSCs) are key players in these processes within vWAT. While second-generation antipsychotics (SGAs) have significantly improved treatments for mental health disorders, their chronic use is associated with an increased risk of MetS. In this study, we explored the impact of SGAs on ADSCs to better understand their role in MetS and identify potential therapeutic targets. Our findings reveal that olanzapine disrupts lipid droplet formation during adipogenic differentiation, impairing insulin receptor endocytosis, turnover, and signaling. SGAs also alter the endolysosomal compartment, leading to acidic vesicle accumulation and increased lysosomal biogenesis through TFEB activation. PKCζ is crucial for the SGA-induced nuclear translocation of TFEB and acidic vesicle formation. Notably, inhibiting PKCζ restored insulin receptor tyrosine phosphorylation, normalized receptor turnover, and improved downstream signaling following olanzapine treatment. This activation of PKCζ by olanzapine is driven by increased phosphatidic acid synthesis via phospholipase D (PLD), following G protein-coupled receptor (GPCR) signaling activation. Overall, olanzapine and clozapine disrupt endolysosomal homeostasis and insulin signaling in a PKCζ-dependent manner. These findings highlight SGAs as valuable tools for uncovering cellular dysfunction in vWAT during MetS and may guide the development of new therapeutic strategies to mitigate the metabolic side effects of these drugs.
Keywords: PKCζ; adipose tissue; adipose-derived mesenchymal stem cells; endocytosis; insulin resistance; insulin signaling; lysosomes; second-generation antipsychotics.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures









References
-
- WHO . WHO Ageing and Health. WHO; Geneve, Switzerland: 2018.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

