DEAD/H Box 5 (DDX5) Augments E2F1-Induced Cell Death Independent of the Tumor Suppressor p53
- PMID: 39769018
- PMCID: PMC11675670
- DOI: 10.3390/ijms252413251
DEAD/H Box 5 (DDX5) Augments E2F1-Induced Cell Death Independent of the Tumor Suppressor p53
Abstract
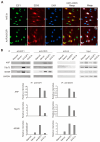
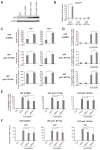
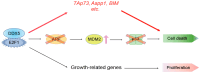
In almost all cancers, the p53 pathway is disabled and cancer cells survive. Hence, it is crucially important to induce cell death independent of p53 in the treatment of cancers. The transcription factor E2F1 is controlled by binding of the tumor suppressor pRB, and induces apoptosis by activating the ARF gene, an upstream activator of p53, when deregulated from pRB by loss of pRB function. Deregulated E2F1 can also induce apoptosis, independent of p53, via other targets such as TAp73 and BIM. We searched for novel E2F1-interacting proteins and identified the RNA helicase DEAD/H box 5 (DDX5), which also functions as a transcriptional coactivator. In contrast to the reported growth-promoting roles of DDX5, we show that DDX5 suppresses cell growth and survival by augmentation of deregulated E2F1 activity. Over-expression of DDX5 enhanced E2F1 induction of tumor suppressor gene expression and cell death. Conversely, shRNA-mediated knockdown of DDX5 compromised both. Moreover, DDX5 modulated E2F1-mediated cell death independent of p53, for which DDX5 also functions as a coactivator. Since p53 function is disabled in almost all cancers, these results underscore the roles of DDX5 in E2F1-mediated induction of cell death, independent of p53, and represent novel aspects for the treatment of p53-disabled cancer cells.
Keywords: ARF; BIM; DDX5; E2F1; TAp73; cell death; p53; pRB.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

