Novel Selectable Marker Sesquiterpenoid Antibiotic Pentalenolactone
- PMID: 39769093
- PMCID: PMC11727764
- DOI: 10.3390/ijms252413328
Novel Selectable Marker Sesquiterpenoid Antibiotic Pentalenolactone
Abstract
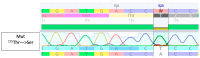
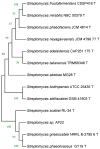
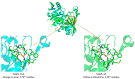
Antibiotic resistance has been and remains a major problem in our society. The main solution to this problem is to search and study the mechanisms of antibiotic action. Many groups of secondary metabolites, including antimicrobial ones, are produced by the Actinomycetota phylum. The actinobacterial strains isolated from habitats that have not been well studied are of great interest. Due to high resource competition, antibiotics are now considered a 'trump card in the game of life' due to their presence in natural substrates with limited nutrients. Potentially, strains isolated from such habitats can be producers of novel or poorly studied antibiotics. In the current research, we identified the strain Streptomyces sp. AP22 from the soils of the Akhshatyrsky Gorge, which is capable of producing pentalenolactone. This study describes the phenotypic and morphological characteristics of Streptomyces sp. AP22 and its biological activity. Pentalenolactone is a known inhibitor of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), an important enzyme involved in glycolysis. We identified a previously unknown mutation in the gapA gene encoding glyceraldehyde-3-phosphate dehydrogenase that confers resistance to this antibiotic compound. This antibiotic is not used in clinical practice, so its application as a selectable marker will not lead to the creation of pathogens resistant to clinically relevant antibiotics. In this case, the selectable marker is based on a genetic construct containing the glyceraldehyde-3-phosphate dehydrogenase gene with a resistance mutation. The use of this selectable marker can be applied to various genetic and molecular techniques, such as cloning and transformation. This can help to facilitate genetic and molecular biology studies of strains resistant to standard selectable markers such as kanamycin or ampicillin.
Keywords: antibiotics; pentalenolactone; selectable marker.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

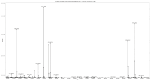

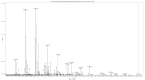

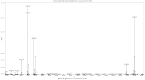
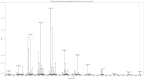









References
-
- Silva G.D.C., Kitano I.T., Ribeiro I.A.D.F., Lacava P.T. The Potential Use of Actinomycetes as Microbial Inoculants and Biopesticides in Agriculture. Front. Soil Sci. 2022;2:833181. doi: 10.3389/fsoil.2022.833181. - DOI
-
- English A.R., Mcbride T.J., Lynch J.E. PA 132, a new antibiotic. II. In vitro and in vivo studies. Antibiot. Annu. 1956:676–681. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

