Should Artificial Intelligence Play a Durable Role in Biomedical Research and Practice?
- PMID: 39769135
- PMCID: PMC11676049
- DOI: 10.3390/ijms252413371
Should Artificial Intelligence Play a Durable Role in Biomedical Research and Practice?
Abstract
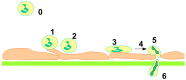
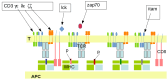
During the last decade, artificial intelligence (AI) was applied to nearly all domains of human activity, including scientific research. It is thus warranted to ask whether AI thinking should be durably involved in biomedical research. This problem was addressed by examining three complementary questions (i) What are the major barriers currently met by biomedical investigators? It is suggested that during the last 2 decades there was a shift towards a growing need to elucidate complex systems, and that this was not sufficiently fulfilled by previously successful methods such as theoretical modeling or computer simulation (ii) What is the potential of AI to meet the aforementioned need? it is suggested that recent AI methods are well-suited to perform classification and prediction tasks on multivariate systems, and possibly help in data interpretation, provided their efficiency is properly validated. (iii) Recent representative results obtained with machine learning suggest that AI efficiency may be comparable to that displayed by human operators. It is concluded that AI should durably play an important role in biomedical practice. Also, as already suggested in other scientific domains such as physics, combining AI with conventional methods might generate further progress and new applications, involving heuristic and data interpretation.
Keywords: deep learning; immunology; machine learning; omic; statistics.
Conflict of interest statement
The author declares no conflict of interest.
Figures
References
-
- Wang Y., Zhang S.-R., Momtaz A., Moradi R., Rastegarnia F., Sahakyan N., Shakeri S., Li L. Can AI Understand Our Universe? Test of Fine-Tuning GPT by Astrophysical Data. arXiv. 20242404.10019
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

