Keratinocyte-Mediated Antigen Presentation in Psoriasis: Preliminary Insights from In Vitro Studies
- PMID: 39769151
- PMCID: PMC11676545
- DOI: 10.3390/ijms252413387
Keratinocyte-Mediated Antigen Presentation in Psoriasis: Preliminary Insights from In Vitro Studies
Abstract
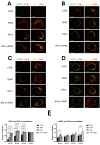
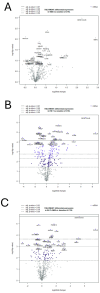
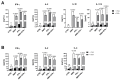
Antigen presentation plays a critical role in the pathogenesis of immune-mediated disorders. This study aimed to investigate the effects of IFN-γ and a cytokine mix (5MIX: IL-1α, IL-17A, IL-22, OsM, and TNF-α) on the antigen-presenting capabilities of keratinocytes, with a specific focus on immune-mediated dermatological conditions such as psoriasis (Ps). To achieve this, keratinocytes were treated with IFN-γ and 5MIX, and their impact on the expression of key antigen-presentation molecules, HLA-DRα and CD74, was assessed. Transcriptomic analysis revealed that IFN-γ alone altered the expression of 254 genes, highlighting its central role in modulating immune responses, including the recruitment of immune cells and regulation of inflammation. Temporal experiments further demonstrated that IFN-γ and 5MIX enhanced early endocytic activity and lysosomal degradation pathways, both essential for effective antigen presentation and T-cell activation. To extend these findings to a clinical context, a co-culture model using keratinocytes derived from psoriatic patients was established. This model revealed increased cytokine production following antigen stimulation, indicating robust and consistent CD4+ and naïve T-cell responses. These results elucidate the complex dynamics of cytokine signaling and antigen presentation in keratinocytes, providing insights into potential therapeutic strategies for immune-mediated skin disorders like Ps.
Keywords: MHC class II; autoimmune diseases; keratinocytes; psoriasis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Overexpression of CD1d by keratinocytes in psoriasis and CD1d-dependent IFN-gamma production by NK-T cells.J Immunol. 2000 Oct 1;165(7):4076-85. doi: 10.4049/jimmunol.165.7.4076. J Immunol. 2000. PMID: 11034419
-
In vitro and in situ expression of IL-23 by keratinocytes in healthy skin and psoriasis lesions: enhanced expression in psoriatic skin.J Immunol. 2006 Feb 1;176(3):1908-15. doi: 10.4049/jimmunol.176.3.1908. J Immunol. 2006. PMID: 16424222
-
Impaired IFN-gamma-dependent inflammatory responses in human keratinocytes overexpressing the suppressor of cytokine signaling 1.J Immunol. 2002 Jul 1;169(1):434-42. doi: 10.4049/jimmunol.169.1.434. J Immunol. 2002. PMID: 12077274
-
Understanding psoriatic disease at single-cell resolution: an update.Curr Opin Rheumatol. 2025 Jul 1;37(4):254-260. doi: 10.1097/BOR.0000000000001085. Epub 2025 Mar 29. Curr Opin Rheumatol. 2025. PMID: 40160177 Review.
-
Resident skin cells in psoriasis: a special look at the pathogenetic functions of keratinocytes.Clin Dermatol. 2007 Nov-Dec;25(6):581-8. doi: 10.1016/j.clindermatol.2007.08.013. Clin Dermatol. 2007. PMID: 18021896 Review.
Cited by
-
IL-15 Promotes the Survival of Anti-Inflammatory (M2), Immunoinhibitory (IL-10+) Dermal Macrophages in Human Eyelid Skin Under IFNγ-Dominated Inflammatory Conditions.Int J Mol Sci. 2025 Aug 13;26(16):7811. doi: 10.3390/ijms26167811. Int J Mol Sci. 2025. PMID: 40869132 Free PMC article.
-
Decoding Non-Coding RNA Regulators in DITRA: From Genomic Insights to Potential Biomarkers and Therapeutic Targets.Genes (Basel). 2025 Jun 27;16(7):753. doi: 10.3390/genes16070753. Genes (Basel). 2025. PMID: 40725409 Free PMC article.
References
-
- Orlik C., Berschneider K.M., Jahraus B., Niesler B., Balta E., Schäkel K., Schröder-Braunstein J., Souto-Carneiro M.M., Samstag Y. Keratinocyte-induced costimulation of human T cells through CD6—but not CD2—activates mTOR and prevents oxidative stress. Front. Immunol. 2022;13:1016112. doi: 10.3389/fimmu.2022.1016112. - DOI - PMC - PubMed
-
- Orlik C., Deibel D., Küblbeck J., Balta E., Ganskih S., Habicht J., Niesler B., Schröder-Braunstein J., Schäkel K., Wabnitz G., et al. Keratinocytes costimulate naive human T cells via CD2: A potential target to prevent the development of proinflammatory Th1 cells in the skin. Cell Mol. Immunol. 2020;17:380–394. doi: 10.1038/s41423-019-0261-x. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

