Multiple Myeloma: Genetic and Epigenetic Biomarkers with Clinical Potential
- PMID: 39769169
- PMCID: PMC11679576
- DOI: 10.3390/ijms252413404
Multiple Myeloma: Genetic and Epigenetic Biomarkers with Clinical Potential
Abstract
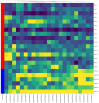
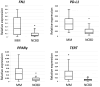
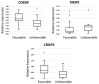
Multiple myeloma (MM) is characterized by the uncontrolled proliferation of monoclonal plasma cells and accounts for approximately 10% of all hematologic malignancies. The clinical outcomes of MM can exhibit considerable variability. Variability in both the genetic and epigenetic characteristics of MM undeniably contributes to tumor dynamics. The aim of the present study was to identify biomarkers with the potential to improve the accuracy of prognosis assessment in MM. Initially, miRNA sequencing was conducted on bone marrow (BM) samples from patients with MM. Subsequently, the expression levels of 27 microRNAs (miRNA) and the gene expression levels of ASF1B, CD82B, CRISP3, FN1, MEF2B, PD-L1, PPARγ, TERT, TIMP1, TOP2A, and TP53 were evaluated via real-time reverse transcription polymerase chain reaction in BM samples from patients with MM exhibiting favorable and unfavorable prognoses. Additionally, the analysis involved the bone marrow samples from patients undergoing examinations for non-cancerous blood diseases (NCBD). The findings indicate a statistically significant increase in the expression levels of miRNA-124, -138, -10a, -126, -143, -146b, -20a, -21, -29b, and let-7a and a decrease in the expression level of miRNA-96 in the MM group compared with NCBD (p < 0.05). No statistically significant differences were detected in the expression levels of the selected miRNAs between the unfavorable and favorable prognoses in MM groups. The expression levels of ASF1B, CD82B, and CRISP3 were significantly decreased, while those of FN1, MEF2B, PDL1, PPARγ, and TERT were significantly increased in the MM group compared to the NCBD group (p < 0.05). The MM group with a favorable prognosis demonstrated a statistically significant decline in TIMP1 expression and a significant increase in CD82B and CRISP3 expression compared to the MM group with an unfavorable prognosis (p < 0.05). From an empirical point of view, we have established that the complex biomarker encompassing the CRISP3/TIMP1 expression ratio holds promise as a prognostic marker in MM. From a fundamental point of view, we have demonstrated that the development of MM is rooted in a cascade of complex molecular pathways, demonstrating the interplay of genetic and epigenetic factors.
Keywords: microRNA; multiple myeloma; oncogenes; prognostic biomarkers; tumor suppressors.
Conflict of interest statement
The authors declare no conflicts of interest. Sergei E. Titov and Mikhail K. Ivanov are Employed by AO Vector-Best Ltd. The company had no role in the design, collection, analysis, or interpretation of data, the writing of the manuscript, or the decision to publish the results.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

