Epigenetic and Cellular Reprogramming of Doxorubicin-Resistant MCF-7 Cells Treated with Curcumin
- PMID: 39769180
- PMCID: PMC11679585
- DOI: 10.3390/ijms252413416
Epigenetic and Cellular Reprogramming of Doxorubicin-Resistant MCF-7 Cells Treated with Curcumin
Abstract
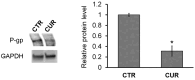
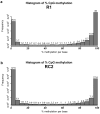
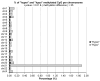
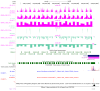
The MCF-7R breast cancer cell line, developed by treating the parental MCF-7 cells with increasing doses of doxorubicin, serves as a model for studying acquired multidrug resistance (MDR). MDR is a major challenge in cancer therapy, often driven by overexpression of the efflux pump P-glycoprotein (P-gp) and epigenetic modifications. While many P-gp inhibitors show promise in vitro, their nonspecific effects on the efflux pump limit in vivo application. Curcumin, a natural compound with pleiotropic action, is a nontoxic P-gp inhibitor capable of modulating multiple pathways. To explore curcumin's molecular effects on MCF-7R cells, we analyzed the expression of genes involved in DNA methylation and transcription regulation, including ABCB1/MDR1. Reduced representation bisulfite sequencing further unveiled key epigenetic changes induced by curcumin. Our findings indicate that curcumin treatment not only modulates critical cellular processes, such as ribosome biogenesis and cytoskeletal dynamics, but also reverses the resistant phenotype, toward that of sensitive cells. This study highlights curcumin's potential as an adjuvant therapy to overcome chemoresistance, offering new avenues for pharmacological strategies targeting epigenetic regulation to re-sensitize resistant cancer cells.
Keywords: DNA methylation; P-glycoprotein; breast cancer; curcumin; cytoskeletal dynamics; multidrug resistance; ribosome biogenesis; translation.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of this study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





Similar articles
-
Curcumin and doxorubicin encapsulated in biocompatible clay-based nanomaterial: A strategy to overcome multidrug resistance.Arch Pharm (Weinheim). 2025 Jan;358(1):e2400702. doi: 10.1002/ardp.202400702. Arch Pharm (Weinheim). 2025. PMID: 39726071 Free PMC article.
-
Forced expression of heat shock protein 27 (Hsp27) reverses P-glycoprotein (ABCB1)-mediated drug efflux and MDR1 gene expression in Adriamycin-resistant human breast cancer cells.J Biol Chem. 2011 Sep 23;286(38):33289-300. doi: 10.1074/jbc.M111.249102. Epub 2011 Jul 22. J Biol Chem. 2011. PMID: 21784846 Free PMC article.
-
Dasatinib reverses the multidrug resistance of breast cancer MCF-7 cells to doxorubicin by downregulating P-gp expression via inhibiting the activation of ERK signaling pathway.Cancer Biol Ther. 2015;16(1):106-14. doi: 10.4161/15384047.2014.987062. Cancer Biol Ther. 2015. PMID: 25482933 Free PMC article.
-
Not only P-glycoprotein: Amplification of the ABCB1-containing chromosome region 7q21 confers multidrug resistance upon cancer cells by coordinated overexpression of an assortment of resistance-related proteins.Drug Resist Updat. 2017 May;32:23-46. doi: 10.1016/j.drup.2017.10.003. Epub 2017 Oct 16. Drug Resist Updat. 2017. PMID: 29145976 Review.
-
Molecular pathways: regulation and therapeutic implications of multidrug resistance.Clin Cancer Res. 2012 Apr 1;18(7):1863-9. doi: 10.1158/1078-0432.CCR-11-1590. Epub 2012 Feb 16. Clin Cancer Res. 2012. PMID: 22344233 Free PMC article. Review.
Cited by
-
Doxorubicin-Induced Cardiotoxicity: A Comprehensive Update.J Cardiovasc Dev Dis. 2025 May 30;12(6):207. doi: 10.3390/jcdd12060207. J Cardiovasc Dev Dis. 2025. PMID: 40558642 Free PMC article. Review.
References
-
- Chekhun V.F., Kulik G.I., Yurchenko O.V., Tryndyak V.P., Todor I.N., Luniv L.S., Tregubova N.A., Pryzimirska T.V., Montgomery B., Rusetskaya N.V., et al. Role of DNA Hypomethylation in the Development of the Resistance to Doxorubicin in Human MCF-7 Breast Adenocarcinoma Cells. Cancer Lett. 2006;231:87–93. doi: 10.1016/j.canlet.2005.01.038. - DOI - PubMed
-
- David G.L., Yegnasubramanian S., Kumar A., Marchi V.L., Marzo A.M.D., Lin X., Nelson W.G. MDR1 Promoter Hypermethylation in MCF-7 Human Breast Cancer Cells: Changes in Chromatin Structure Induced by Treatment with 5-Aza-Cytidine. Cancer Biol. Ther. 2004;3:540–548. doi: 10.4161/cbt.3.6.845. - DOI - PubMed
-
- Ai L., Kim W.-J., Demircan B., Dyer L.M., Bray K.J., Skehan R.R., Massoll N.A., Brown K.D. The Transglutaminase 2 Gene (TGM2), a Potential Molecular Marker for Chemotherapeutic Drug Sensitivity, Is Epigenetically Silenced in Breast Cancer. Carcinogenesis. 2008;29:510–518. doi: 10.1093/carcin/bgm280. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

