Functional and Biological Characterization of the LGR5Δ5 Splice Variant in HEK293T Cells
- PMID: 39769183
- PMCID: PMC11678308
- DOI: 10.3390/ijms252413417
Functional and Biological Characterization of the LGR5Δ5 Splice Variant in HEK293T Cells
Abstract
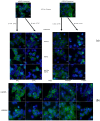
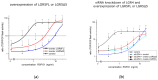
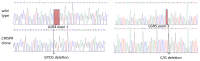
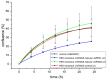
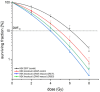
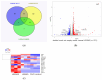
The regulator of the canonical Wnt pathway, leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5), is expressed in the stem cell compartment of several tissues and overexpressed in different human carcinomas. The isoform of the stem cell marker LGR5, named LGR5Δ5 and first described by our group, is associated with prognosis and metastasis in oral squamous cell carcinoma (OSCC) and soft tissue sarcoma (STS). In a proof-of-principle analysis, the function of LGR5Δ5 was investigated in HEK293T cells, a model cell line of the Wnt pathway, compared to full-length LGR5 (FL) expression. The CRISPR/CAS knockout of LGR5 and LGR4 (thereby avoiding the side effects of LGR4) resulted in a loss of Wnt activity that cannot be restored by LGR5Δ5 but by LGR5FL rescue. The ability to migrate was not affected by LGR5Δ5, but was reduced by LGR5FL overexpression. The CRISPR/CAS of LGR4 and 5 induced radiosensitization, which was enhanced by the overexpression of LGR5FL or LGR5Δ5. RNA sequencing analysis revealed a significant increase in the ligand R-spondin 1 (RSPO1) level by LGR5Δ5. Furthermore, LGR5Δ5 appears to be involved in the regulation of genes related to the cytoskeleton, extracellular matrix stiffness, and angiogenesis, while LGR5FL is associated with the regulation of collagens and histone proteins.
Keywords: CRISPR/CAS; LGR4; LGR5; LGR5Δ5; isoform; stem cell marker.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Ghobakhloo S., Khoshhali M., Vatandoost N., Jafarpour S., Niazmand A., Nedaeinia R., Salehi R. Clinical Implications and Prognostic Value of Leucine-Rich G Protein-Coupled Receptor 5 Expression as A Cancer Stem Cell Marker in Malignancies: A Systematic Review and Meta-Analysis. Cell J. 2024;26:1–12. doi: 10.22074/CELLJ.2023.2010157.1396. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

