MHTAPred-SS: A Highly Targeted Autoencoder-Driven Deep Multi-Task Learning Framework for Accurate Protein Secondary Structure Prediction
- PMID: 39769208
- PMCID: PMC11677681
- DOI: 10.3390/ijms252413444
MHTAPred-SS: A Highly Targeted Autoencoder-Driven Deep Multi-Task Learning Framework for Accurate Protein Secondary Structure Prediction
Abstract
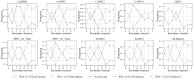
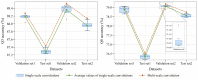
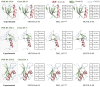
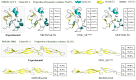
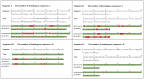
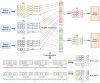
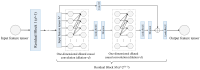
Accurate protein secondary structure prediction (PSSP) plays a crucial role in biopharmaceutics and disease diagnosis. Current prediction methods are mainly based on multiple sequence alignment (MSA) encoding and collaborative operations of diverse networks. However, existing encoding approaches lead to poor feature space utilization, and encoding quality decreases with fewer homologous proteins. Moreover, the performance of simple stacked networks is greatly limited by feature extraction capabilities and learning strategies. To this end, we propose MHTAPred-SS, a novel PSSP framework based on the fusion of six features, including the embedding feature derived from a pre-trained protein language model. First, we propose a highly targeted autoencoder (HTA) as the driver to encode sequences in a homologous protein-independent manner. Second, under the guidance of biological knowledge, we design a protein secondary structure prediction model based on the multi-task learning strategy (PSSP-MTL). Experimental results on six independent test sets show that MHTAPred-SS achieves state-of-the-art performance, with values of 88.14%, 84.89%, 78.74% and 77.15% for Q3, SOV3, Q8 and SOV8 metrics on the TEST2016 dataset, respectively. Additionally, we demonstrate that MHTAPred-SS has significant advantages in single-category and boundary secondary structure prediction, and can finely capture the distribution of secondary structure segments, thereby contributing to subsequent tasks.
Keywords: deep multi-task learning; highly targeted autoencoder; multi-feature fusion; pre-trained protein language model; protein secondary structure prediction.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

