Improved Genetic Characterization of Hypercholesterolemia in Latvian Patients with Familial Hypercholesterolemia: A Combined Monogenic and Polygenic Approach Using Whole-Genome Sequencing
- PMID: 39769230
- PMCID: PMC11677843
- DOI: 10.3390/ijms252413466
Improved Genetic Characterization of Hypercholesterolemia in Latvian Patients with Familial Hypercholesterolemia: A Combined Monogenic and Polygenic Approach Using Whole-Genome Sequencing
Abstract
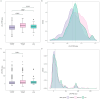
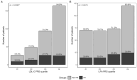
Despite the implementation of next-generation sequencing-based genetic testing on patients with clinical familial hypercholesterolemia (FH), most cases lack complete genetic characterization. We aim to investigate the utility of the polygenic risk score (PRS) in specifying the genetic background of patients from the Latvian Registry of FH (LRFH). We analyzed the whole-genome sequencing (WGS) data of the clinically diagnosed FH patients (n = 339) and controls selected from the Latvian reference population (n = 515). Variant pathogenicity in FH patients was classified according to the ACMG/AMP guidelines. The low-density lipoprotein cholesterol (LDL-C) and lipoprotein (a) (LPA) PRS were calculated based on the WGS data. We identified unique causative variants in 80 (23.6%) of the tested individuals (39 variants in FH genes and 4 variants in phenocopy genes, with 6 variants being novel). The LDL-C PRS was highly discriminative compared to the LPA PRS. Nevertheless, both PRS were able to explain the genetic cause of hypercholesterolemia in 26.3% of the remaining non-monogenic patients. The combined genetic analysis of monogenic and polygenic hypercholesterolemia resulted in 43.7% genetically explained hypercholesterolemia cases. Even though the application of PRS alone does not exclude monogenic testing in clinical FH patients, it is a valuable tool for diagnosis specification.
Keywords: familial hypercholesterolemia; lipoprotein (a); low-density lipoprotein cholesterol; monogenic hypercholesterolemia; polygenic risk score; severe hypercholesterolemia; whole-genome sequencing.
Conflict of interest statement
G.N. has attended conferences sponsored by Abbott, Amgen, Biotronik, and Medtronic. V.S. has given talks and attended conferences sponsored by Amgen, Sanofi, and Servier Laboratories. D.G. has attended conferences sponsored by Amgen, Sanofi, Novartis, Servier, and Pfizer. R.M. has given talks and attended conferences sponsored by Amgen, Bayer, Sanofi, Servier Laboratories, Swixx BioPharma, Cardurion Pharmaceuticals, and 89bio. E.T. has attended conferences sponsored by Amgen, Swixx BioPharma, and NovoNordisk. G.L. has given talks, attended conferences, and received consultancy fees from Abbott Laboratories, Amgen, Astra-Zeneca, Bayer, Boehringer Ingelheim, GlaxoSmithKline, Grindex, KRKA, MSD, Mylan, Novartis, Novo Nordisk, Pfizer, Roche Laboratories, Sanofi, Servier Laboratories, Siemens Laboratories, and SwixxBiopharma, Zentiva.
Figures
References
-
- Cuchel M., Bruckert E., Ginsberg H.N., Raal F.J., Santos R.D., Hegele R.A., Kuivenhoven J.A., Nordestgaard B.G., Descamps O.S., Steinhagen-Thiessen E., et al. Homozygous Familial Hypercholesterolaemia: New Insights and Guidance for Clinicians to Improve Detection and Clinical Management. A Position Paper from the Consensus Panel on Familial Hypercholesterolaemia of the European Atherosclerosis Society. Eur. Heart J. 2014;35:2146–2157. doi: 10.1093/eurheartj/ehu274. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

