Molecular Insights into Structural Dynamics and Binding Interactions of Selected Inhibitors Targeting SARS-CoV-2 Main Protease
- PMID: 39769245
- PMCID: PMC11677904
- DOI: 10.3390/ijms252413482
Molecular Insights into Structural Dynamics and Binding Interactions of Selected Inhibitors Targeting SARS-CoV-2 Main Protease
Abstract
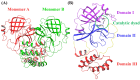
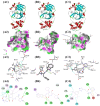
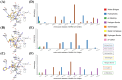
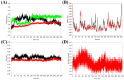
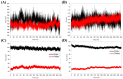
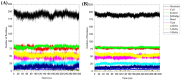
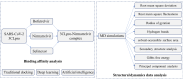
The SARS-CoV-2 main protease (Mpro, also known as 3CLpro) is a key target for antiviral therapy due to its critical role in viral replication and maturation. This study investigated the inhibitory effects of Bofutrelvir, Nirmatrelvir, and Selinexor on 3CLpro through molecular docking, molecular dynamics (MD) simulations, and free energy calculations. Nirmatrelvir exhibited the strongest binding affinity across docking tools (AutoDock Vina: -8.3 kcal/mol; DiffDock: -7.75 kcal/mol; DynamicBound: 7.59 to 7.89 kcal/mol), outperforming Selinexor and Bofutrelvir. Triplicate 300 ns MD simulations revealed that the Nirmatrelvir-3CLpro complex displayed high conformational stability, reduced root mean square deviation (RMSD), and a modest decrease in solvent-accessible surface area (SASA), indicating enhanced structural rigidity. Gibbs free energy analysis highlighted greater flexibility in unbound 3CLpro, stabilized by Nirmatrelvir binding, supported by stable hydrogen bonds. MolProphet prediction tools, targeting the Cys145 residue, confirmed that Nirmatrelvir exhibited the strongest binding, forming multiple hydrophobic, hydrogen, and π-stacking interactions with key residues, and had the lowest predicted IC50/EC50 (9.18 × 10-8 mol/L), indicating its superior potency. Bofutrelvir and Selinexor showed weaker interactions and higher IC50/EC50 values. MM/PBSA analysis calculated a binding free energy of -100.664 ± 0.691 kJ/mol for the Nirmatrelvir-3CLpro complex, further supporting its stability and binding potency. These results underscore Nirmatrelvir's potential as a promising therapeutic agent for SARS-CoV-2 and provide novel insights into dynamic stabilizing interactions through AI-based docking and long-term MD simulations.
Keywords: Bofutrelvir; Paxlovid; SARS-CoV-2; Selinexor; artificial intelligence; molecular interactions.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Singh M., Jayant K., Singh D., Bhutani S., Poddar N.K., Chaudhary A.A., Khan S.U., Adnan M., Siddiqui A.J., Hassan M.I., et al. Withania somnifera (L.) Dunal (Ashwagandha) for the possible therapeutics and clinical management of SARS-CoV-2 infection: Plant-based drug discovery and targeted therapy. Front. Cell Infect. Microbiol. 2022;12:933824. doi: 10.3389/fcimb.2022.933824. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

