Metabolomic and Lipidomic Profiling for Pre-Transplant Assessment of Delayed Graft Function Risk Using Chemical Biopsy with Microextraction Probes
- PMID: 39769265
- PMCID: PMC11728147
- DOI: 10.3390/ijms252413502
Metabolomic and Lipidomic Profiling for Pre-Transplant Assessment of Delayed Graft Function Risk Using Chemical Biopsy with Microextraction Probes
Abstract
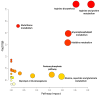
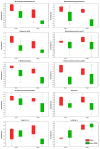
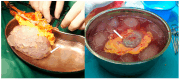
Organ shortage remains a significant challenge in transplantology, prompting efforts to maximize the use of available organs and expand the donor pool, including through extended criteria donors (ECDs). However, ECD kidney recipients often face poorer outcomes, including a higher incidence of delayed graft function (DGF), which is linked to worse graft performance, reduced long-term survival, and an increased need for interventions like dialysis. This underscores the urgent need for strategies to improve early DGF risk assessment and optimize post-transplant management for high-risk patients. This study conducted multi-time point metabolomic and lipidomic analyses of donor kidney tissue and recipient plasma to identify compounds predicting DGF risk and assess the translational potential of solid-phase microextraction (SPME) for graft evaluation and early complication detection. The SPME-based chemical biopsy enabled a direct kidney analysis, while thin-film microextraction facilitated high-throughput plasma preparation. Following high-performance liquid chromatography coupled with a mass spectrometry analysis, the random forest algorithm was applied to identify compounds with predictive potential for assessing DGF risk before transplantation. Additionally, a comparison of metabolomic and lipidomic profiles of recipient plasma during the early post-operative days identified metabolites that distinguish between DGF and non-DGF patients. The selected compounds primarily included amino acids and their derivatives, nucleotides, organic acids, peptides, and lipids, particularly phospholipids and triacylglycerols. In conclusion, this study highlights the significant translational potential of chemical biopsies and plasma metabolite analyses for risk assessments and the non-invasive monitoring of DGF. The identified metabolites provide a foundation for developing a comprehensive DGF assessment and monitoring method, with potential integration into routine clinical practice.
Keywords: DGF; LC-MS; graft quality assessment; kidney transplantation; lipidomics; metabolomics; solid-phase microextraction.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Pre-transplant HLA Antibodies and Delayed Graft Function in the Current Era of Kidney Transplantation.Front Immunol. 2020 Aug 26;11:1886. doi: 10.3389/fimmu.2020.01886. eCollection 2020. Front Immunol. 2020. PMID: 32983110 Free PMC article.
-
Identification of expanded-criteria donor kidney grafts at lower risk of delayed graft function.Transplantation. 2013 Oct 15;96(7):633-8. doi: 10.1097/TP.0b013e31829d9225. Transplantation. 2013. PMID: 23912171
-
Metabolomic and lipidomic landscape of porcine kidney associated with kidney perfusion in heart beating donors and donors after cardiac death.Transl Res. 2024 May;267:79-90. doi: 10.1016/j.trsl.2023.12.001. Epub 2023 Dec 3. Transl Res. 2024. PMID: 38052298
-
Machine perfusion preservation versus static cold storage for deceased donor kidney transplantation.Cochrane Database Syst Rev. 2019 Mar 15;3(3):CD011671. doi: 10.1002/14651858.CD011671.pub2. Cochrane Database Syst Rev. 2019. Update in: Cochrane Database Syst Rev. 2024 Jul 9;7:CD011671. doi: 10.1002/14651858.CD011671.pub3. PMID: 30875082 Free PMC article. Updated.
-
Risk factors of delayed graft function following living donor kidney transplantation: A meta-analysis.Transpl Immunol. 2024 Oct;86:102094. doi: 10.1016/j.trim.2024.102094. Epub 2024 Jul 23. Transpl Immunol. 2024. PMID: 39053613
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

