Exploring Protein Functions of Gut Bacteriome and Mycobiome in Thai Infants Associated with Atopic Dermatitis Through Metaproteomic and Host Interaction Analysis
- PMID: 39769296
- PMCID: PMC11676981
- DOI: 10.3390/ijms252413533
Exploring Protein Functions of Gut Bacteriome and Mycobiome in Thai Infants Associated with Atopic Dermatitis Through Metaproteomic and Host Interaction Analysis
Abstract
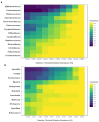
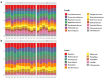
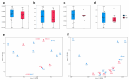
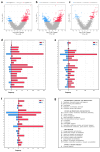
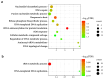
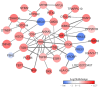
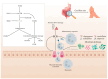
Atopic dermatitis (AD), a prevalent allergic skin condition in children, has been closely associated with imbalances in the gut microbiome. To investigate these microbial alterations and their functional implications, we investigated protein expression, functions and interactions of the gut bacteriome and mycobiome as well as the human proteome in Thai infants with AD using integrative metaproteomic and host interaction analysis. As we observed, probiotic species, such as Lactobacillus acidophilus and Bacteroides salyersiae, were reduced in abundance in the AD group while key pathogenic bacteria and fungi, such as Streptococcus constellatus and Penicillium chrysogenum, increased in abundance. Additionally, the functional analysis of expressed proteins was enriched in response to stress and DNA repair in the bacteriome and ribosome biogenesis-related processes in the mycobiome of the AD group, potentially associated to increased reactive oxygen species (ROS), intestinal inflammation, fungal growth and microbial dysbiosis. Further, a protein-protein interactions (PPIs) network analysis incorporating the human proteome revealed 10 signature proteins related to stress and immune system processes associated with AD. Our findings propose the interactions of the key species and signature protein functions between the gut microbes and the human host in response to AD in Thai infants. To our knowledge, this study serves as the first framework for monitoring bacteriome-mycobiome-human gut studies associated with AD and other allergic diseases in infants.
Keywords: atopic dermatitis; human gut; metaproteomics; microbiome; mycobiome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Archer C.B. Atopic dermatitis. Medicine. 2021;49:370–373. doi: 10.1016/j.mpmed.2021.03.006. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

