H3K4me3 Genome-Wide Distribution and Transcriptional Regulation of Transposable Elements by RNA Pol2 Deposition
- PMID: 39769308
- PMCID: PMC11677803
- DOI: 10.3390/ijms252413545
H3K4me3 Genome-Wide Distribution and Transcriptional Regulation of Transposable Elements by RNA Pol2 Deposition
Abstract
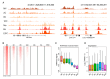
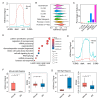
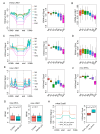
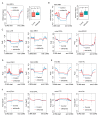
Zygotic genome activation (ZGA) is critical for early embryo development and is meticulously regulated by epigenetic modifications. H3K4me3 is a transcription-permissive histone mark preferentially found at promoters, but its distribution across genome features remains incompletely understood. In this study, we investigated the genome-wide enrichment of H3K4me3 during early embryo development and embryonic stem cells (ESCs) in both sheep and mice. We discovered that broad H3K4me3 domains were present in MII stage oocytes and were progressively diminished, while promoter H3K4me3 enrichment was increased and correlated with gene upregulation during ZGA in sheep. Additionally, we reported the dynamic distribution of H3K4me3 at the transposable elements (TEs) during early embryo development in both sheep and mice. Specifically, the H3K4me3 distribution of LINE1 and ERVL, two subsets of TEs, was associated with their expression during early embryo development in sheep. Furthermore, H3K4me3 enrichment in TEs was greatly increased during ZGA following Kdm5b knockdown, and the distribution of RNA polymerase II (Pol2) in TEs was also markedly increased in Kdm5b knockout ESCs in mice. These findings suggest that H3K4me3 plays important roles in regulating TE expression through interaction with RNA Pol2, providing valuable insights into the regulation of ZGA initiation and cell fate determination by H3K4me3.
Keywords: H3K4me3; RNA Pol2; embryonic stem cells; transposable elements; zygotic genome activation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Sotomayor-Lugo F., Iglesias-Barrameda N., Castillo-Aleman Y.M., Casado-Hernandez I., Villegas-Valverde C.A., Bencomo-Hernandez A.A., Ventura-Carmenate Y., Rivero-Jimenez R.A. The Dynamics of Histone Modifications during Mammalian Zygotic Genome Activation. Int. J. Mol. Sci. 2024;25:1459. doi: 10.3390/ijms25031459. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

