BCAT1 Associates with DNA Repair Proteins KU70 and KU80 and Contributes to Regulate DNA Repair in T-Cell Acute Lymphoblastic Leukemia (T-ALL)
- PMID: 39769333
- PMCID: PMC11676169
- DOI: 10.3390/ijms252413571
BCAT1 Associates with DNA Repair Proteins KU70 and KU80 and Contributes to Regulate DNA Repair in T-Cell Acute Lymphoblastic Leukemia (T-ALL)
Abstract
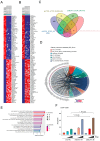
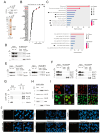
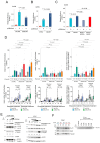
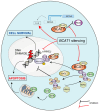
Increased expression of branched-chain amino acid (BCAA) transaminase 1 (BCAT1) often correlates with tumor aggressiveness and drug resistance in cancer. We have recently reported that BCAT1 was overexpressed in a subgroup of T-cell acute lymphoblastic (T-ALL) samples, especially those with NOTCH1 activating mutations. Interestingly, BCAT1-depleted cells showed pronounced sensitivity to DNA-damaging agents such as etoposide; however, how BCAT1 regulates this sensitivity remains uncertain. Here, we provide further clues on its chemo-sensitizing effect. Indeed, BCAT1 protein regulates the non-homologous end joining (c-NHEJ) DNA repair pathway by physically associating with the KU70/KU80 heterodimer. BCAT1 inhibition during active repair of DNA double-strand breaks (DSBs) led to increased KU70/KU80 acetylation and impaired c-NHEJ repair, a dramatic increase in DSBs, and ultimately cell death. Our results suggest that, in T-ALL, BCAT1 possesses non-metabolic functions that confer a drug resistance mechanism and that targeting BCAT1 activity presents a novel strategy to improve chemotherapy response in T-ALL patients.
Keywords: BCAT1; T-cell lymphoblastic leukemia; leukemia growth; metabolism.
Conflict of interest statement
Author Adonia E. Papathanassiu was employed by the company Ergon Pharmaceuticals, LLC. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

