Beyond Nucleotide Excision Repair: The Importance of XPF in Base Excision Repair and Its Impact on Cancer, Inflammation, and Aging
- PMID: 39769376
- PMCID: PMC11728164
- DOI: 10.3390/ijms252413616
Beyond Nucleotide Excision Repair: The Importance of XPF in Base Excision Repair and Its Impact on Cancer, Inflammation, and Aging
Abstract
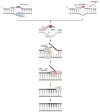
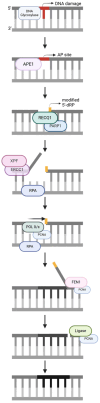
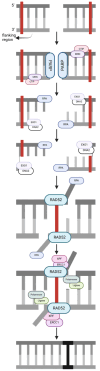
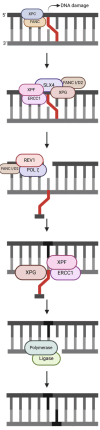
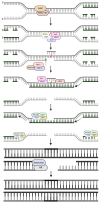
DNA repair involves various intricate pathways that work together to maintain genome integrity. XPF (ERCC4) is a structural endonuclease that forms a heterodimer with ERCC1 that is critical in both single-strand break repair (SSBR) and double-strand break repair (DSBR). Although the mechanistic function of ERCC1/XPF has been established in nucleotide excision repair (NER), its role in long-patch base excision repair (BER) has recently been discovered through the 5'-Gap pathway. This study briefly explores the roles of XPF in different pathways to emphasize the importance of XPF in DNA repair. XPF deficiency manifests in various diseases, including cancer, neurodegeneration, and aging-related disorders; it is also associated with conditions such as Xeroderma pigmentosum and fertility issues. By examining the molecular mechanisms and pathological consequences linked to XPF dysfunction, this study aims to elucidate the crucial role of XPF in genomic stability as a repair protein in BER and provide perspectives regarding its potential as a therapeutic target in related diseases.
Keywords: 5′-Gap pathway (5′ Gap LP-BER pathway); RECQ1; XPF (ERCC4); base excision repair (BER); nucleotide excision repair (NER).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
The ERCC1 and ERCC4 (XPF) genes and gene products.Gene. 2015 Sep 15;569(2):153-61. doi: 10.1016/j.gene.2015.06.026. Epub 2015 Jun 12. Gene. 2015. PMID: 26074087 Free PMC article. Review.
-
CARM1/PRMT4 facilitates XPF-ERCC1 heterodimer assembly and maintains nucleotide excision repair activity.Nucleic Acids Res. 2025 Apr 22;53(8):gkaf355. doi: 10.1093/nar/gkaf355. Nucleic Acids Res. 2025. PMID: 40304182 Free PMC article.
-
Multiple roles of the ERCC1-XPF endonuclease in DNA repair and resistance to anticancer drugs.Anticancer Res. 2010 Sep;30(9):3223-32. Anticancer Res. 2010. PMID: 20944091 Review.
-
Mislocalization of XPF-ERCC1 nuclease contributes to reduced DNA repair in XP-F patients.PLoS Genet. 2010 Mar 5;6(3):e1000871. doi: 10.1371/journal.pgen.1000871. PLoS Genet. 2010. PMID: 20221251 Free PMC article.
-
Interstrand crosslink repair: can XPF-ERCC1 be let off the hook?Trends Genet. 2008 Feb;24(2):70-6. doi: 10.1016/j.tig.2007.11.003. Epub 2008 Jan 14. Trends Genet. 2008. PMID: 18192062 Review.
Cited by
-
Associations Between DNA Repair Gene Polymorphisms and Breast Cancer Histopathological Subtypes: A Preliminary Study.J Clin Med. 2025 May 27;14(11):3764. doi: 10.3390/jcm14113764. J Clin Med. 2025. PMID: 40507526 Free PMC article.
-
Comprehensive review on Fanconi anemia: insights into DNA interstrand cross-links, repair pathways, and associated tumors.Orphanet J Rare Dis. 2025 Jul 30;20(1):389. doi: 10.1186/s13023-025-03896-w. Orphanet J Rare Dis. 2025. PMID: 40739565 Free PMC article. Review.
References
-
- Sijbers A.M., de Laat W.L., Ariza R.R., Biggerstaff M., Wei Y.F., Moggs J.G., Carter K.C., Shell B.K., Evans E., de Jong M.C., et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/S0092-8674(00)80155-5. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

