Tertiary Structures of Haseki Tick Virus Nonstructural Proteins Are Similar to Those of Orthoflaviviruses
- PMID: 39769413
- PMCID: PMC11678601
- DOI: 10.3390/ijms252413654
Tertiary Structures of Haseki Tick Virus Nonstructural Proteins Are Similar to Those of Orthoflaviviruses
Abstract
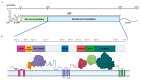
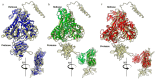
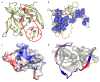
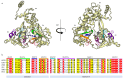
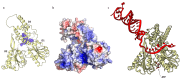
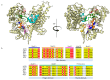
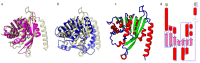
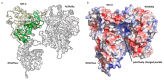
Currently, a large number of novel tick-borne viruses potentially pathogenic to humans are discovered. Studying many of them by classical methods of virology is difficult due to the absence of live viral particles or a sufficient amount of their genetic material. In this case, the use of modern methods of bioinformatics and synthetic and structural biology can help. Haseki tick virus (HSTV) is a recently discovered tick-borne unclassified ssRNA(+) virus. HSTV-positive patients experienced fever and an elevated temperature. However, at the moment, there is no information on the tertiary structure and functions of its proteins. In this work, we used AlphaFold 3 and other bioinformatic tools for the annotation of HSTV nonstructural proteins, based on the principle that the tertiary structure of a protein is inextricably linked with its molecular function. We were the first to obtain models of tertiary structures and describe the putative functions of HSTV nonstructural proteins (NS3 helicase, NS3 protease, NS5 RNA-dependent RNA-polymerase, and NS5 methyltransferase), which play a key role in viral genome replication. Our results may help in further taxonomic identification of HSTV and the development of direct-acting antiviral drugs, POC tests, and vaccines.
Keywords: AlphaFold 3; Flaviviridae; RNA viruses; ixodid ticks; orthoflavi-like viruses; protein structure; tick-borne infection; viral proteins.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Zerbini F.M., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Adriaenssens E.M., Alfenas-Zerbini P., Dempsey D.M., Dutilh B.E., García M.L., Hendrickson R.C., et al. Changes to Virus Taxonomy and the ICTV Statutes Ratified by the International Committee on Taxonomy of Viruses (2023) Arch. Virol. 2023;168:175. doi: 10.1007/s00705-023-05797-4. - DOI - PMC - PubMed
-
- Paul Chrystal . The History of the World in 100 Pandemic. Pen and Sword History; Barnsley, UK: 2021.
-
- Ergunay K., Bourke B.P., Reinbold-Wasson D.D., Nikolich M.P., Nelson S.P., Caicedo-Quiroga L., Vaydayko N., Kirkitadze G., Chunashvili T., Long L.S., et al. The Expanding Range of Emerging Tick-Borne Viruses in Eastern Europe and the Black Sea Region. Sci. Rep. 2023;13:19824. doi: 10.1038/s41598-023-46879-2. - DOI - PMC - PubMed
-
- Shi M., Lin X.-D., Vasilakis N., Tian J.-H., Li C.-X., Chen L.-J., Eastwood G., Diao X.-N., Chen M.-H., Chen X., et al. Divergent Viruses Discovered in Arthropods and Vertebrates Revise the Evolutionary History of the Flaviviridae and Related Viruses. J. Virol. 2016;90:659–669. doi: 10.1128/JVI.02036-15. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

