Characterization of Site-Specific N- and O-Glycopeptides from Recombinant Spike and ACE2 Glycoproteins Using LC-MS/MS Analysis
- PMID: 39769415
- PMCID: PMC11678118
- DOI: 10.3390/ijms252413649
Characterization of Site-Specific N- and O-Glycopeptides from Recombinant Spike and ACE2 Glycoproteins Using LC-MS/MS Analysis
Abstract
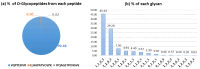
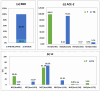
The COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in hundreds of millions of infections and millions of deaths globally. Although vaccination campaigns are mitigating the pandemic, emerging viral variants continue to pose challenges. The spike (S) protein of SARS-CoV-2 plays a critical role in viral entry by binding to the angiotensin-converting enzyme 2 (ACE2) receptor, making both proteins essential targets for therapeutic and vaccine development. The glycosylation of these proteins influences their structure and function. This underscores the need for detailed site-specific glycoproteomic analysis. In this study, we characterized the N- or O-glycosylation profiles of the recombinant receptor-binding domain (RBD) of spike protein and ACE2 proteins expressed from Expi293F cells, as well as the S2 subunit of spike protein expressed in plant (N. benthamiana) cells. Using a high-resolution Orbitrap Eclipse Tribrid mass spectrometer equipped with the Ultimate 3000 RSLCnano and I-GPA (Integrated GlycoProteome Analyzer) developed in a previous study, 148 N- and 28 O-glycopeptides from RBD, 71 N-glycopeptides from the S2 subunit, and 139 N-glycopeptides from ACE2 were characterized. In addition, we report post-translational modifications (PTMs) of glycan, including mannose-6-phosphate (M6P) and GlcNAc-1-phosphate-6-O-mannose in N-glycan of RBD and ACE2, and O-acetylation in O-glycan of RBD, identified for the first time in these recombinant proteins. The relative abundance distribution according to glycosites and glycan types were analyzed by quantified site-specific N- and O (only from RBD)-glycopeptides from RBD, S2, and ACE2 using I-GPA. Asn331 for RBD, Asn1098 for S2, and Asn103 for ACE2 were majorly N-glycosylated, and dominant glycan-type was complex from RBD and ACE2 and high-mannose from S2. These findings will provide valuable insights into the glycosylation patterns that influence protein function and immunogenicity and offer new perspectives for the development of vaccines and antibody-based therapies against COVID-19.
Keywords: LC-MS/MS analysis; modification of glycan; reaction-binding domain; recombinant proteins; site-specific glycopeptides; spike glycoprotein.
Conflict of interest statement
The authors declare the following financial interest/personal relationships which may be considered as potentially competing.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

