Somatic Instability Leading to Mosaicism in Fragile X Syndrome and Associated Disorders: Complex Mechanisms, Diagnostics, and Clinical Relevance
- PMID: 39769443
- PMCID: PMC11728179
- DOI: 10.3390/ijms252413681
Somatic Instability Leading to Mosaicism in Fragile X Syndrome and Associated Disorders: Complex Mechanisms, Diagnostics, and Clinical Relevance
Abstract
Fragile X syndrome (FXS) is a genetic condition caused by the inheritance of alleles with >200 CGG repeats in the 5' UTR of the fragile X messenger ribonucleoprotein 1 (FMR1) gene. These full mutation (FM) alleles are associated with DNA methylation and gene silencing, which result in intellectual disabilities, developmental delays, and social and behavioral issues. Mosaicism for both the size of the CGG repeat tract and the extent of its methylation is commonly observed in individuals with the FM. Mosaicism has also been reported in carriers of premutation (PM) alleles, which have 55-200 CGG repeats. PM alleles confer risk for the fragile X premutation-associated conditions (FXPAC), including FXTAS, FXPOI, and FXAND, conditions thought to be due to the toxic consequences of transcripts containing large CGG-tracts. Unmethylated FM (UFM) alleles are transcriptionally and translationally active. Thus, they produce transcripts with toxic effects. These transcripts do produce some FMRP, the encoded product of the FMR1 gene, albeit with reduced translational efficiency. As a result, mosaicism can result in a complex clinical presentation. Here, we review the concept of mosaicism in both FXS and in PM carriers, including its potential clinical significance.
Keywords: FMR1 gene; activation ratio; full mutation; methylation; mosaicism; premutation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

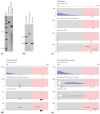
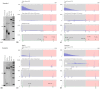
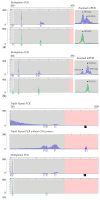
References
-
- Wright G.E.B., Collins J.A., Kay C., McDonald C., Dolzhenko E., Xia Q., Bečanović K., Drögemöller B.I., Semaka A., Nguyen C.M., et al. Length of Uninterrupted CAG, Independent of Polyglutamine Size, Results in Increased Somatic Instability, Hastening Onset of Huntington Disease. Am. J. Hum. Genet. 2019;104:1116–1126. doi: 10.1016/j.ajhg.2019.04.007. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

