Genomic Analysis and Functional Validation of Bidirectional Promoters in Medaka (Oryzias latipes)
- PMID: 39769487
- PMCID: PMC11676430
- DOI: 10.3390/ijms252413726
Genomic Analysis and Functional Validation of Bidirectional Promoters in Medaka (Oryzias latipes)
Abstract
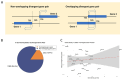
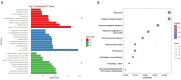
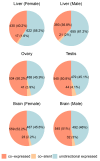
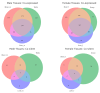
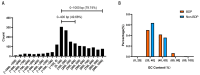
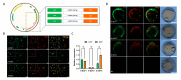
Bidirectional promoters (BDPs) regulate the transcription of two adjacent, oppositely oriented genes, offering a compact structure with significant potential for multigene expression systems. Although BDPs are evolutionarily conserved, their regulatory roles and sequence characteristics vary across species, with limited studies in fish. Here, we systematically analyzed the distribution, sequence features, and expression patterns of BDPs in the medaka (Oryzias latipes) genome. A total of 1737 divergent gene pairs, representing 13% of medaka genes, were identified as potentially regulated by BDPs. These genes are enriched in essential biological processes, including organelle function, RNA processing, and ribosome biogenesis. Transcriptomic analysis revealed that co-regulation (co-expression and co-silencing) is a prominent feature of these gene pairs, with variability influenced by tissue and sex. Sequence analysis showed that medaka BDPs are compact, with most fragments under 400 bp and an average GC content of 42.06%. Validation experiments confirmed the bidirectional transcriptional activity of three histone-related BDPs in both medaka SG3 cells and embryos, demonstrating effective and robust regulatory efficiency. This study enhances our understanding of the genomic organization and transcriptional regulation in fish and provides a valuable reference for developing species-specific multigene expression systems in fish genetic engineering.
Keywords: bidirectional promoters; gene regulation; medaka; multigene expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

