Transposable Elements Contribute to the Regulation of Long Noncoding RNAs in Drosophila melanogaster
- PMID: 39769552
- PMCID: PMC11678190
- DOI: 10.3390/insects15120950
Transposable Elements Contribute to the Regulation of Long Noncoding RNAs in Drosophila melanogaster
Abstract
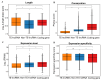
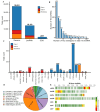
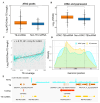
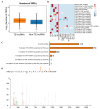
Background: Transposable elements (TEs) and noncoding sequences are major components of the genome, yet their functional contributions to long noncoding RNAs (lncRNAs) are not well understood. Although many lncRNAs originating from TEs (TE-lncRNAs) have been identified across various organisms, their characteristics and regulatory roles, particularly in insects, remain largely unexplored. This study integrated multi-omics data to investigate TE-lncRNAs in D. melanogaster, focusing on the influence of transposons across different omics levels. Results: We identified 16,118 transposons overlapping with lncRNA sequences that constitute 2119 TE-lncRNAs (40.4% of all lncRNAs) using 256 public RNA-seq samples and 15 lncRNA-seq samples of Drosophila S2 cells treated with heavy metals. Of these, 67.2% of TE-lncRNAs contain more than one TE. The LTR/Gypsy family was the most common transposon insertion. Transposons preferred to insert into promoters, transcription starting sites, and intronic regions, especially in chromosome ends. Compared with lncRNAs, TE-lncRNAs showed longer lengths, a lower conservation, and lower levels but a higher specificity of expression. Multi-omics data analysis revealed positive correlations between transposon insertions and chromatin openness at the pre-transcriptional level. Notably, a total of 516 TE-lncRNAs provided transcriptional factor binding sites through transposon insertions. The regulatory network of a key transcription factor was rewired by transposons, potentially recruiting other transcription factors to exert regulatory functions under heavy metal stress. Additionally, 99 TE-lncRNAs were associated with m6A methylation modification sites, and 115 TE-lncRNAs potentially provided candidate small open reading frames through transposon insertions. Conclusions: Our data analysis demonstrated that TEs contribute to the regulation of lncRNAs. TEs not only promote the transcriptional regulation of lncRNAs, but also facilitate their post-transcriptional and epigenetic regulation.
Keywords: Drosophila; TE-lncRNA; heavy metal; long noncoding RNA; transposable element.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Stamatoyannopoulos J.A., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. - DOI - PMC - PubMed
-
- Liu Y., Shi M., He X., Cao Y., Liu P., Li F., Zou S., Wen C., Zhan Q., Xu Z., et al. LncRNA-PACERR induces pro-tumour macrophages via interacting with miR-671-3p and m6A-reader IGF2BP2 in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 2022;15:52. doi: 10.1186/s13045-022-01272-w. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

