Targeting mTOR Kinase with Natural Compounds: Potent ATP-Competitive Inhibition Through Enhanced Binding Mechanisms
- PMID: 39770519
- PMCID: PMC11677242
- DOI: 10.3390/ph17121677
Targeting mTOR Kinase with Natural Compounds: Potent ATP-Competitive Inhibition Through Enhanced Binding Mechanisms
Abstract
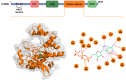
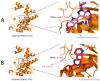
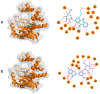
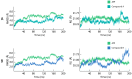
Background/Objectives: The mammalian target of the rapamycin (mTOR) signaling pathway is a central regulator of cell growth, proliferation, metabolism, and survival. Dysregulation of mTOR signaling contributes to many human diseases, including cancer, diabetes, and obesity. Therefore, inhibitors against mTOR's catalytic kinase domain (KD) have been developed and have shown significant antitumor activities, making it a promising therapeutic target. The ATP-KD interaction is particularly important for mTOR to exert its cellular functions, and such inhibitors have demonstrated efficient attenuation of overall mTOR activity. Methods: In this study, we screened the Traditional Chinese Medicine (TCM) database, which enlists natural products that capture the relationships between drugs targets and diseases. Our aim was to identify potential ATP-competitive agonists that target the mTOR-KD and compete with ATP to bind the mTOR-KD serving as potential potent mTOR inhibitors. Results: We identified two compounds that demonstrated interatomic interactions similar to those of ATP-mTOR. The conformational stability and dynamic features of the mTOR-KD bound to the selected compounds were tested by subjecting each complex to 200 ns molecular dynamic (MD) simulations and molecular mechanics/generalized Born surface area (MM/GBSA) to extract free binding energies. We show the effectiveness of both compounds in forming stable complexes with the mTOR-KD, which is more effective than the mTOR-KD-ATP complex with more robust binding affinities. Conclusions: This study implies that both compounds could serve as potential therapeutic inhibitors of mTOR, regulating its function and, therefore, mitigating human disease progression.
Keywords: ATP; kinase domain; mTOR; mTOR inhibitors; molecular simulation; natural compounds.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
New Mammalian Target of Rapamycin (mTOR) Modulators Derived from Natural Product Databases and Marine Extracts by Using Molecular Docking Techniques.Mar Drugs. 2018 Oct 15;16(10):385. doi: 10.3390/md16100385. Mar Drugs. 2018. PMID: 30326670 Free PMC article.
-
Marine-Derived Natural Products as ATP-Competitive mTOR Kinase Inhibitors for Cancer Therapeutics.Pharmaceuticals (Basel). 2021 Mar 21;14(3):282. doi: 10.3390/ph14030282. Pharmaceuticals (Basel). 2021. PMID: 33801030 Free PMC article.
-
Antitumor activities of ATP-competitive inhibitors of mTOR in colon cancer cells.BMC Cancer. 2012 Mar 8;12:86. doi: 10.1186/1471-2407-12-86. BMC Cancer. 2012. PMID: 22401294 Free PMC article.
-
New inhibitors of the PI3K-Akt-mTOR pathway: insights into mTOR signaling from a new generation of Tor Kinase Domain Inhibitors (TORKinibs).Curr Top Microbiol Immunol. 2010;347:241-62. doi: 10.1007/82_2010_64. Curr Top Microbiol Immunol. 2010. PMID: 20549474 Review.
-
The mTOR Signaling Pathway: Key Regulator and Therapeutic Target for Heart Disease.Biomedicines. 2025 Feb 7;13(2):397. doi: 10.3390/biomedicines13020397. Biomedicines. 2025. PMID: 40002810 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

