The Small RNA MicF Represses ObgE and SeqA in Escherichia coli
- PMID: 39770600
- PMCID: PMC11676804
- DOI: 10.3390/microorganisms12122397
The Small RNA MicF Represses ObgE and SeqA in Escherichia coli
Abstract
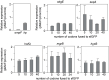
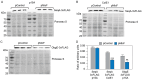
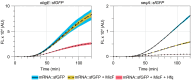
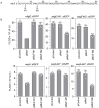
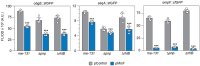
Small regulatory RNAs (sRNA) have been shown to play a large role in the management of stress responses in Escherichia coli and other bacteria. Upon fluctuations in nutrient availability and exposure to antimicrobials and superoxide-generating agents, the MicF sRNA in E. coli has been shown to regulate a small set of genes involved in the management of membrane permeability. Currently, it is unknown whether MicF acts on other processes to mediate the response to these agents. Using an sRNA interaction prediction tool, we identified genes in E. coli that are potentially regulated by MicF. Through subsequent analysis using a sfGFP-based reporter-gene fusion, we have validated two novel targets of MicF regulation: ObgE, a GTPase crucial for chromosome partitioning, and SeqA, a negative modulator of DNA replication. Importantly, the interaction between MicF and these target mRNAs is contingent upon the presence of the RNA chaperone protein, Hfq. Furthermore, our findings affirm the role of MicF's conserved 5' seed pairing region in initiating these regulatory interactions. Our study suggests that, beyond its established role in membrane permeability management, MicF exerts control over chromosome dynamics in response to distinct environmental cues, implicating a more multifaceted regulatory function in bacterial stress adaptation.
Keywords: Escherichia coli; RNA; gene regulation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Update of
-
Beyond membrane permeability: A role for the small RNA MicF in regulation of chromosome replication and partitioning.bioRxiv [Preprint]. 2024 Apr 22:2024.04.22.590647. doi: 10.1101/2024.04.22.590647. bioRxiv. 2024. Update in: Microorganisms. 2024 Nov 22;12(12):2397. doi: 10.3390/microorganisms12122397. PMID: 38712278 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources

