Metagenome-Assembled Genomes of Pig Fecal Samples in Nine European Countries: Insights into Antibiotic Resistance Genes and Viruses
- PMID: 39770612
- PMCID: PMC11676251
- DOI: 10.3390/microorganisms12122409
Metagenome-Assembled Genomes of Pig Fecal Samples in Nine European Countries: Insights into Antibiotic Resistance Genes and Viruses
Abstract
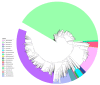
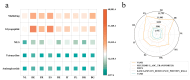
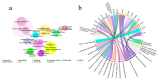
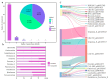
Gut microbiota plays a crucial role in the health and productivity of pigs. However, the spread of antibiotic resistance genes (ARGs) and viruses within the pig intestinal microbiota poses significant threats to animal and public health. This study utilized 181 pig samples from nine European countries and employed metagenomic assembly methods to investigate the dynamics and distribution of ARGs and viruses within the pig intestinal microbiota, aiming to observing their associations with potential bacterial hosts. We identified 4605 metagenome-assembled genomes (MAGs), corresponding to 19 bacterial phyla, 97 families, 309 genera, and a total of 449 species. Additionally, 44 MAGs were classified as archaea. Analysis of ARGs revealed 276 ARG types across 21 ARG classes, with Glycopeptide being the most abundant ARG class, followed by the class of Multidrug. Treponema D sp016293915 was identified as a primary potential bacterial host for Glycopeptide. Aligning nucleotide sequences with a viral database, we identified 1044 viruses. Among the viral genome families, Peduoviridae and Intestiviridae were the most prevalent, with CAG-914 sp000437895 being the most common potential host species for both. These findings highlight the importance of MAGs in enhancing our understanding of the gut microbiome, revealing microbial diversity, antibiotic resistance, and virus-bacteria interactions. The data analysis for the article was based on the public dataset PRJEB22062 in the European Nucleotide Archive.
Keywords: antibiotic resistance genes; metagenome-assembled genomes; pig gut; viruses.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Genome-centric analyses of 165 metagenomes show that mobile genetic elements are crucial for the transmission of antimicrobial resistance genes to pathogens in activated sludge and wastewater.Microbiol Spectr. 2024 Mar 5;12(3):e0291823. doi: 10.1128/spectrum.02918-23. Epub 2024 Jan 30. Microbiol Spectr. 2024. PMID: 38289113 Free PMC article.
-
Deciphering the mobility and bacterial hosts of antibiotic resistance genes under antibiotic selection pressure by metagenomic assembly and binning approaches.Water Res. 2020 Nov 1;186:116318. doi: 10.1016/j.watres.2020.116318. Epub 2020 Aug 25. Water Res. 2020. PMID: 32871290
-
Novel Insights into the Pig Gut Microbiome Using Metagenome-Assembled Genomes.Microbiol Spectr. 2022 Aug 31;10(4):e0238022. doi: 10.1128/spectrum.02380-22. Epub 2022 Jul 26. Microbiol Spectr. 2022. PMID: 35880887 Free PMC article.
-
Metagenome-assembled genomes and gene catalog from the chicken gut microbiome aid in deciphering antibiotic resistomes.Commun Biol. 2021 Nov 18;4(1):1305. doi: 10.1038/s42003-021-02827-2. Commun Biol. 2021. PMID: 34795385 Free PMC article.
-
Profiles of Microbial Community and Antibiotic Resistome in Wild Tick Species.mSystems. 2022 Aug 30;7(4):e0003722. doi: 10.1128/msystems.00037-22. Epub 2022 Aug 1. mSystems. 2022. PMID: 35913190 Free PMC article. Review.
Cited by
-
Unlocking the Potential of Metagenomics with the PacBio High-Fidelity Sequencing Technology.Microorganisms. 2024 Dec 2;12(12):2482. doi: 10.3390/microorganisms12122482. Microorganisms. 2024. PMID: 39770685 Free PMC article. Review.
-
Unraveling the composition and function of pig gut microbiome from metagenomics.Anim Microbiome. 2025 Jun 4;7(1):60. doi: 10.1186/s42523-025-00419-7. Anim Microbiome. 2025. PMID: 40468444 Free PMC article. Review.
References
-
- Ricker N., Trachsel J., Colgan P., Jones J., Choi J., Lee J., Coetzee J.F., Howe A., Brockmeier S.L., Loving C.L. Toward antibiotic stewardship: Route of antibiotic administration impacts the microbiota and resistance gene diversity in swine feces. Front. Vet. Sci. 2020;7:255. doi: 10.3389/fvets.2020.00255. - DOI - PMC - PubMed
-
- European Centre for Disease Prevention and Control . Antimicrobial Resistance Surveillance in Europe 2015. ECDC; Stockholm, Sweden: 2015. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net)
Grants and funding
LinkOut - more resources
Full Text Sources

