Whole-Genome Sequencing of Peribacillus frigoritolerans Strain d21.2 Isolated in the Republic of Dagestan, Russia
- PMID: 39770615
- PMCID: PMC11678259
- DOI: 10.3390/microorganisms12122410
Whole-Genome Sequencing of Peribacillus frigoritolerans Strain d21.2 Isolated in the Republic of Dagestan, Russia
Abstract
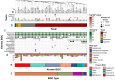
Pesticide-free agriculture is a fundamental pillar of environmentally friendly agriculture. To this end, there is an active search for new bacterial strains capable of synthesizing secondary metabolites and toxins that protect crops from pathogens and pests. In this study, we isolated a novel strain d21.2 of Peribacillus frigoritolerans from a soil sample collected in the Republic of Dagestan, Russia. Leveraging several bioinformatic approaches on Illumina-based whole-genome assembly, we revealed that the strain harbors certain insecticidal loci (coding for putative homologs of Bmp and Vpa) and also contains multiple BGCs (biosynthetic gene clusters), including paeninodin, koranimine, schizokinen, and fengycin. In total, 21 BGCs were predicted as synthesizing metabolites with bactericidal and/or fungicidal effects. Importantly, by applying a re-scaffolding pipeline, we managed to robustly predict MGEs (mobile genetic elements) associated with BGCs, implying high genetic plasticity. In addition, the d21.2's genome was free from genes encoding for enteric toxins, implying its safety in use. A comparison with available genomes of the Peribacillus frigoritolerans strain revealed that the strain described here contains more functionally important loci than other members of the species. Therefore, strain d21.2 holds potential for use in agriculture due to the probable manifestation of bactericidal, fungicidal, growth-stimulating, and other useful properties. The assembled genome is available in the NCBI GeneBank under ASM4106054v1.
Keywords: Illumina sequencing; Peribacillus frigoritolerans; bactericidal activity; biosynthetic gene clusters; draft genome; fungicidal properties; mobile genetic elements.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Plant growth-promoting rhizobacteria: Peribacillus frigoritolerans 2RO30 and Pseudomonas sivasensis 2RO45 for their effect on canola growth under controlled as well as natural conditions.Front Plant Sci. 2024 Jan 8;14:1233237. doi: 10.3389/fpls.2023.1233237. eCollection 2023. Front Plant Sci. 2024. PMID: 38259930 Free PMC article.
-
Whole-Genome Sequencing and Biotechnological Potential Assessment of Two Bacterial Strains Isolated from Poultry Farms in Belgorod, Russia.Microorganisms. 2023 Sep 5;11(9):2235. doi: 10.3390/microorganisms11092235. Microorganisms. 2023. PMID: 37764079 Free PMC article.
-
Genomic Insights into the Bactericidal and Fungicidal Potential of Bacillus mycoides b12.3 Isolated in the Soil of Olkhon Island in Lake Baikal, Russia.Microorganisms. 2024 Nov 28;12(12):2450. doi: 10.3390/microorganisms12122450. Microorganisms. 2024. PMID: 39770653 Free PMC article.
-
Reclassification of Brevibacterium frigoritolerans as Peribacillus frigoritolerans comb. nov. based on phylogenomics and multiple molecular synapomorphies.Int J Syst Evol Microbiol. 2022 May;72(5). doi: 10.1099/ijsem.0.005389. Int J Syst Evol Microbiol. 2022. PMID: 35604831
-
Simply Versatile: The Use of Peribacillus simplex in Sustainable Agriculture.Microorganisms. 2023 Oct 12;11(10):2540. doi: 10.3390/microorganisms11102540. Microorganisms. 2023. PMID: 37894197 Free PMC article. Review.
References
-
- Świątczak J., Kalwasińska A., Brzezinska M.S. Plant Growth–Promoting Rhizobacteria: Peribacillus frigoritolerans 2RO30 and Pseudomonas sivasensis 2RO45 for Their Effect on Canola Growth under Controlled as Well as Natural Conditions. Front. Plant Sci. 2024;14:1233237. doi: 10.3389/fpls.2023.1233237. - DOI - PMC - PubMed
-
- Selvakumar G., Sushil S.N., Stanley J., Mohan M., Deol A., Rai D., Ramkewal, Bhatt J.C., Gupta H.S. Brevibacterium frigoritolerans a Novel Entomopathogen of Anomala dimidiata and Holotrichia longipennis (Scarabaeidae: Coleoptera) Biocontrol Sci. Technol. 2011;21:821–827. doi: 10.1080/09583157.2011.586021. - DOI
-
- Marik D., Sharma P., Chauhan N.S., Jangir N., Shekhawat R.S., Verma D., Mukherjee M., Abiala M., Roy C., Yadav P., et al. Peribacillus frigoritolerans T7-IITJ, a Potential Biofertilizer, Induces Plant Growth-Promoting Genes of Arabidopsis thaliana. J. Appl. Microbiol. 2024;135:lxae066. doi: 10.1093/jambio/lxae066. - DOI - PubMed
-
- Chacón-López A., Guardado-Valdivia L., Bañuelos-González M., López-García U., Montalvo-González E., Arvizu-Gómez J., Stoll A., Aguilera S. Effect of Metabolites Produced by Bacillus atrophaeus and Brevibacterium frigoritolerans Strains on Postharvest Biocontrol of Alternaria alternata in Tomato (Solanum lycopersicum L.) Biocontrol Sci. 2021;26:67–74. doi: 10.4265/bio.26.67. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

