Exploring the Linkage Between Ruminal Microbial Communities on Postweaning and Finishing Diets and Their Relation to Residual Feed Intake in Beef Cattle
- PMID: 39770639
- PMCID: PMC11676184
- DOI: 10.3390/microorganisms12122437
Exploring the Linkage Between Ruminal Microbial Communities on Postweaning and Finishing Diets and Their Relation to Residual Feed Intake in Beef Cattle
Abstract
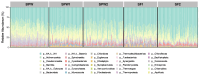
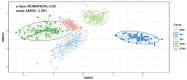
Feed efficiency significantly impacts the economics of beef production and is influenced by biological and environmental factors. The rumen microbiota plays a crucial role in efficiency, with studies increasingly focused on its relationship with different rearing systems. This study analyzed 324 rumen samples from bulls and steers categorized as high and low efficiency based on residual feed intake. The animals were fed two diets (postweaning and finishing) and rumen samples were sequenced using a reduced representation sequencing (RRS) based approach. The results indicated that diet significantly affected microbial diversity and abundance. In postweaning diets, Actinomycetota, particularly Bifidobacterium, were prevalent, aiding carbohydrate fermentation. In contrast, Acetoanaerobium was identified in finishing diets, likely contributing to acetate production. Additionally, Bacteroides and Butyrivibrio were abundant during postweaning, known for fiber degradation and volatile fatty acid production. Notably, Prevotella and Fibrobacter succinogenes were associated with high feed intake and nutrient utilization, indicating their potential as microbial biomarkers. However, alpha diversity indices showed no significant relationship with feed efficiency, suggesting that diversity alone may not adequately reflect the complexity of feed efficiency phenotypes. These findings highlight the importance of diet and microbial interactions on feed efficiency and suggest further research to explore these microbial contributions to precision feeding strategies.
Keywords: Bos taurus; feed efficiency; microbiome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Effects of lasalocid, narasin, or virginiamycin supplementation on rumen parameters and performance of beef cattle fed forage-based diet.J Anim Sci. 2023 Jan 3;101:skad108. doi: 10.1093/jas/skad108. J Anim Sci. 2023. PMID: 37042805 Free PMC article.
-
Effects of hempseed cake on ruminal fermentation parameters, nutrient digestibility, nutrient flow, and nitrogen balance in finishing steers.J Anim Sci. 2023 Jan 3;101:skac291. doi: 10.1093/jas/skac291. J Anim Sci. 2023. PMID: 36592747 Free PMC article.
-
Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle.Appl Environ Microbiol. 2012 Jul;78(14):4949-58. doi: 10.1128/AEM.07759-11. Epub 2012 May 4. Appl Environ Microbiol. 2012. PMID: 22562991 Free PMC article.
-
Effects of cashew nutshell extract inclusion into a high-grain finishing diet on methane emissions, nutrient digestibility, and ruminal fermentation in beef steers.J Anim Sci. 2025 Jan 4;103:skae359. doi: 10.1093/jas/skae359. J Anim Sci. 2025. PMID: 39574184
-
Effects of bacterial direct-fed microbial mixtures offered to beef cattle consuming finishing diets on intake, nutrient digestibility, feeding behavior, and ruminal kinetics/fermentation profile.J Anim Sci. 2024 Jan 3;102:skae003. doi: 10.1093/jas/skae003. J Anim Sci. 2024. PMID: 38183669 Free PMC article.
References
-
- Koch R.M., Swiger L.A., Chambers D., Gregory K.E. Efficiency of feed use in beef cattle. J. Anim. Sci. 1963;22:486–494. doi: 10.2527/jas1963.222486x. - DOI
-
- Pravia M.I., Navajas E.A., Aguilar I., Ravagnolo O. Evaluation of feed efficiency traits in different Hereford populations and their effect on variance component estimation. Anim. Prod. Sci. 2022;62:1652–1660. doi: 10.1071/AN21420. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

