Genomic Characterization of Bacillus sp. THPS1: A Hot Spring-Derived Species with Functional Features and Biotechnological Potential
- PMID: 39770679
- PMCID: PMC11727782
- DOI: 10.3390/microorganisms12122476
Genomic Characterization of Bacillus sp. THPS1: A Hot Spring-Derived Species with Functional Features and Biotechnological Potential
Abstract
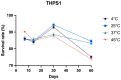
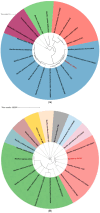
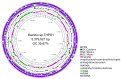
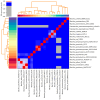
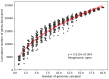
Bacillus sp. THPS1 is a novel strain isolated from a high-temperature hot spring in Thailand, exhibiting distinctive genomic features that enable adaptation to an extreme environment. This study aimed to characterize the genomic and functional attributes of Bacillus sp. THPS1 to understand its adaptation strategies and evaluate its potential for biotechnological applications. The draft genome is 5.38 Mbp with a GC content of 35.67%, encoding 5606 genes, including those linked to stress response and sporulation, which are essential for survival in high-temperature conditions. Phylogenetic analysis and average nucleotide identity (ANI) values confirmed its classification as a distinct species within the Bacillus genus. Pangenome analysis involving 19 others closely related thermophilic Bacillus species identified 1888 singleton genes associated with heat resistance, sporulation, and specialized metabolism, suggesting adaptation to nutrient-deficient, high-temperature environments. Genomic analysis revealed 12 biosynthetic gene clusters (BGCs), including those for polyketides and non-ribosomal peptides, highlighting its potential for synthesizing secondary metabolites that may facilitate its adaptation. Additionally, the presence of three Siphoviridae phage regions and 96 mobile genetic elements (MGEs) suggests significant genomic plasticity, whereas the existence of five CRISPR arrays implies an advanced defense mechanism against phage infections, contributing to genomic stability. The distinctive genomic features and functional capacities of Bacillus sp. THPS1 make it a promising candidate for biotechnological applications, particularly in the production of heat-stable enzymes and the development of resilient bioformulations.
Keywords: Bacillus sp.; aquaculture; genomic adaptation; pangenome analysis; sporulation genes.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Minnaard J., Rolny I.S., Pérez P.F. Laboratory Models for Foodborne Infections. CRC Press; Boca Raton, FL, USA: 2017. Bacillus; pp. 131–154.
-
- Kuan K.B., Othman R., Abdul Rahim K., Shamsuddin Z.H. Plant growth-promoting rhizobacteria inoculation to enhance vegetative growth, nitrogen fixation and nitrogen remobilisation of maize under greenhouse conditions. PLoS ONE. 2016;11:e0152478. doi: 10.1371/journal.pone.0152478. - DOI - PMC - PubMed
-
- Prashar P., Kapoor N., Sachdeva S. Rhizosphere: Its structure, bacterial diversity and significance. Rev. Environ. Sci. Bio/Technol. 2014;13:63–77. doi: 10.1007/s11157-013-9317-z. - DOI
-
- Federici B. Microbiology Monographs Inclusions in Prokaryotes. Springer; Berlin/Heidelberg, Germany: 2006. Insecticidal protein crystals of Bacillus thuringiensis.
LinkOut - more resources
Full Text Sources
Miscellaneous

