Microbial Community Structure, Diversity, and Succession During Decomposition of Kiwifruit Litters with Different Qualities
- PMID: 39770701
- PMCID: PMC11727838
- DOI: 10.3390/microorganisms12122498
Microbial Community Structure, Diversity, and Succession During Decomposition of Kiwifruit Litters with Different Qualities
Abstract
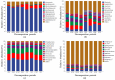
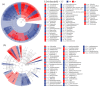
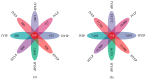
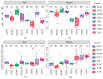
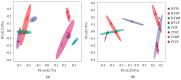
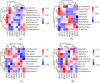
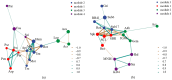
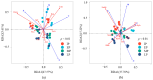
There are differences in the litter quality and decomposition rate of kiwifruit varieties, but it is not clear whether these differences are related to microbial communities. The leaf litters of two kiwifruit varieties (A. chinensis cv 'Hongyang' and A. chinensis cv 'Jinyan') were taken as objects, and the structure, diversity, and succession of the soil microbial communities were analyzed using an in situ decomposition experiment. Moreover, the contents of C, N, P, and K in the litters during decomposition were analyzed. The results show that there were variety differences in community structure at the generic level. Lophotrichus, Acaulium, and Fusarium were relatively more abundant in the microbial community of the 'Hongyang' kiwifruit litter, and Humicola and Tausonia were relatively more abundant in the microbial community of the 'Jinyan' kiwifruit litter. Subgroup_6 and Sphingomonas were the dominant bacteria. The bacterial community diversity of 'Jinyan' kiwifruit was higher than that of the 'Hongyang' kiwifruit litter. The community diversity was higher in the middle and later periods. The contents of C and N in the litters were the main factors affecting microbial communities. The abundances of Humicola and Apiotrichum were negatively correlated with the contents of C and N, and the abundances of Sphingomonas and SC-I-84 were positively correlated with the content of C. There were variety differences in the microbial communities corresponding to the decomposition processes of the 'Hongyang' and 'Jinyan' kiwifruit litters. The mechanisms of the variety differences were related to litter quality and the initial soil microbial community.
Keywords: kiwifruit; litter decomposition; litter quality; microbial community; orchard ecosystem; variety difference.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Krishna M.P., Mohan M. Litter decomposition in forest ecosystems: A review. Energy Ecol. Environ. 2017;2:236–249. doi: 10.1007/s40974-017-0064-9. - DOI
-
- Giweta M. Role of litter production and its decomposition, and factors affecting the processes in a tropical forest ecosystem: A review. J. Ecol. Environ. 2020;44:11. doi: 10.1186/s41610-020-0151-2. - DOI
-
- Wang Q., Chai Q., Dou X., Zhao C., Yin W., Li H., Wei J. Soil Microorganisms in Agricultural Fields and Agronomic Regulation Pathways. Agronomy. 2024;14:669. doi: 10.3390/agronomy14040669. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

