Wild-Mouse-Derived Gut Microbiome Transplantation in Laboratory Mice Partly Alleviates House-Dust-Mite-Induced Allergic Airway Inflammation
- PMID: 39770703
- PMCID: PMC11728220
- DOI: 10.3390/microorganisms12122499
Wild-Mouse-Derived Gut Microbiome Transplantation in Laboratory Mice Partly Alleviates House-Dust-Mite-Induced Allergic Airway Inflammation
Abstract
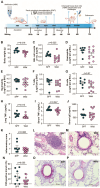
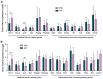
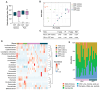
Laboratory mice are instrumental for preclinical research but there are serious concerns that the use of a clean standardized environment for specific-pathogen-free (SPF) mice results in poor bench-to-bedside translation due to their immature immune system. The aim of the present study was to test the importance of the gut microbiota in wild vs. SPF mice for evaluating host immune responses in a house-dust-mite-induced allergic airway inflammation model without the influence of pathogens. The wild mouse microbiome reduced histopathological changes and TNF-α in the lungs and serum when transplanted to microbiota-depleted mice compared to mice transplanted with the microbiome from SPF mice. Moreover, the colonic gene expression of Gata3 was significantly lower in the wild microbiome-associated mice, whereas Muc1 was more highly expressed in both the ileum and colon. Intestinal microbiome and metabolomic analyses revealed distinct profiles associated with the wild-derived microbiome. The wild-mouse microbiome thus partly reduced sensitivity to house-dust-mite-induced allergic airway inflammation compared to the SPF mouse microbiome, and preclinical studies using this model should consider using both 'dirty' rewilded and SPF mice for testing new therapeutic compounds due to the significant effects of their respective microbiomes and derived metabolites on host immune responses.
Keywords: animal models; fecal microbiota transplantation; feral mice; gut microbiota; immunity; microbial metabolites; translatability; wild mice.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
N-(3-oxododecanoyl)-homoserine lactone disrupts intestinal barrier and induces systemic inflammation through perturbing gut microbiome in mice.Sci Total Environ. 2021 Jul 15;778:146347. doi: 10.1016/j.scitotenv.2021.146347. Epub 2021 Mar 11. Sci Total Environ. 2021. PMID: 34030388
-
Meta-omics profiling of the gut-lung axis illuminates metabolic networks and host-microbial interactions associated with elevated lung elastance in a murine model of obese allergic asthma.Front Microbiomes. 2023;2:1153691. doi: 10.3389/frmbi.2023.1153691. Epub 2023 May 5. Front Microbiomes. 2023. PMID: 37293566 Free PMC article.
-
Cross-talk between airway and gut microbiome links to IgE responses to house dust mites in childhood airway allergies.Sci Rep. 2020 Aug 10;10(1):13449. doi: 10.1038/s41598-020-70528-7. Sci Rep. 2020. PMID: 32778700 Free PMC article.
-
House dust microbiome and human health risks.Int Microbiol. 2019 Sep;22(3):297-304. doi: 10.1007/s10123-019-00057-5. Epub 2019 Jan 14. Int Microbiol. 2019. PMID: 30811000 Review.
-
Go With Your Gut: The Shaping of T-Cell Response by Gut Microbiota in Allergic Asthma.Front Immunol. 2020 Jul 14;11:1485. doi: 10.3389/fimmu.2020.01485. eCollection 2020. Front Immunol. 2020. PMID: 32760404 Free PMC article. Review.
Cited by
-
Assessing gut microbial provisioning of essential amino acids to host in a murine model with reconstituted gut microbiomes.Res Sq [Preprint]. 2025 Mar 24:rs.3.rs-6255159. doi: 10.21203/rs.3.rs-6255159/v1. Res Sq. 2025. PMID: 40195995 Free PMC article. Preprint.
References
-
- Rosshart S.P., Vassallo B.G., Angeletti D., Hutchinson D.S., Morgan A.P., Takeda K., Hickman H.D., McCulloch J.A., Badger J.H., Ajami N.J., et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell. 2017;171:1015–1028.e1013. doi: 10.1016/j.cell.2017.09.016. - DOI - PMC - PubMed
-
- Beura L.K., Hamilton S.E., Bi K., Schenkel J.M., Odumade O.A., Casey K.A., Thompson E.A., Fraser K.A., Rosato P.C., Filali-Mouhim A., et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532:512–516. doi: 10.1038/nature17655. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

