Metagenomic Insight into the Associated Microbiome in Plasmodia of Myxomycetes
- PMID: 39770743
- PMCID: PMC11677963
- DOI: 10.3390/microorganisms12122540
Metagenomic Insight into the Associated Microbiome in Plasmodia of Myxomycetes
Abstract
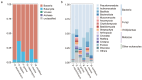
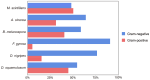
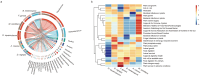
During the trophic period of myxomycetes, the plasmodia of myxomycetes can perform crawling feeding and phagocytosis of bacteria, fungi, and organic matter. Culture-based studies have suggested that plasmodia are associated with one or several species of bacteria; however, by amplicon sequencing, it was shown that up to 31-52 bacteria species could be detected in one myxomycete, suggesting that the bacterial diversity associated with myxomycetes was likely to be underestimated. To fill this gap and characterize myxomycetes' microbiota and functional traits, the diversity and functional characteristics of microbiota associated with the plasmodia of six myxomycetes species were investigated by metagenomic sequencing. The results indicate that the plasmodia harbored diverse microbial communities, including eukaryotes, viruses, archaea, and the dominant bacteria. The associated microbiomes represented more than 22.27% of the plasmodia genome, suggesting that these microbes may not merely be parasitic or present as food but rather may play functional roles within the plasmodium. The six myxomycetes contained similar bacteria, but the bacteria community compositions in each myxomycete were species-specific. Functional analysis revealed a highly conserved microbial functional profile across the six plasmodia, suggesting they may serve a specific function for the myxomycetes. While the host-specific selection may shape the microbial community compositions within plasmodia, functional redundancy ensures functional stability across different myxomycetes.
Keywords: coexistence; core microbiome; functional redundancy; metagenome; slime mold.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Diversity of bacterial communities in the plasmodia of myxomycetes.BMC Microbiol. 2022 Dec 22;22(1):314. doi: 10.1186/s12866-022-02725-5. BMC Microbiol. 2022. PMID: 36544088 Free PMC article.
-
Observations on an ATP-sensitive protein system from the plasmodia of a myxomycete.J Gen Physiol. 1956 Jan 20;39(3):325-47. doi: 10.1085/jgp.39.3.325. J Gen Physiol. 1956. PMID: 13286452 Free PMC article.
-
A simple rapid procedure for obtaining axenic cultures from monoxenic cultures of myxomycete plasmodia.Can J Microbiol. 1999 Oct;45(10):865-70. doi: 10.1139/w99-080. Can J Microbiol. 1999. PMID: 10907423
-
The Species Problem in Myxomycetes Revisited.Protist. 2016 Aug;167(4):319-338. doi: 10.1016/j.protis.2016.05.003. Epub 2016 Jun 6. Protist. 2016. PMID: 27351595 Review.
-
Isolation of bioactive natural products from myxomycetes.Med Chem. 2005 Nov;1(6):575-90. doi: 10.2174/157340605774598135. Med Chem. 2005. PMID: 16787341 Review.
References
-
- Keller H.W., Everhart S.E., Kilgore C.M. Introduction, Basic Biology, Life Cycles, Genetics, and Reproduction. In: Rojas C., Stephenson S.L., editors. Myxomycetes (Biology, Systematics, Biogeography and Ecology) 2nd ed. Elsevier Inc.; Maryland Heights, MO, USA: 2021.
-
- Geisen S., Mitchell E.A.D., Wilkinson D.M., Adl S., Bonkowski M., Brown M.W., Fiore-Donno A.M., Heger T.J., Jassey V.E.J., Krashevska V., et al. Soil Protistology Rebooted: 30 Fundamental Questions to Start With. Soil Biol. Biochem. 2017;111:94–103. doi: 10.1016/j.soilbio.2017.04.001. - DOI
-
- Everhart S.E., Keller H.W. Life History Strategies of Corticolous Myxomycetes: The Life Cycle, Plasmodial Types, Fruiting Bodies, and Taxonomic Orders. Fungal Divers. 2008;29:1–16.
-
- Cohen A.L. Nutrition of the Myxomycetes. II. Relations between Plasmodia, Bacteria, and Substrate in Two-Membered Culture. Bot. Gaz. 1941;103:205–224. doi: 10.1086/335038. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

