UV-C Exposure Enhanced the Cd2+ Adsorption Capability of the Radiation-Resistant Strain Sphingomonas sp. M1-B02
- PMID: 39770822
- PMCID: PMC11678681
- DOI: 10.3390/microorganisms12122620
UV-C Exposure Enhanced the Cd2+ Adsorption Capability of the Radiation-Resistant Strain Sphingomonas sp. M1-B02
Abstract
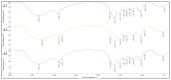
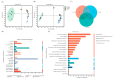
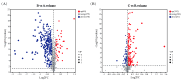
Microbial adsorption is a cost-effective and environmentally friendly remediation method for heavy metal pollution. The adsorption mechanism of cadmium (Cd) by bacteria inhabiting extreme environments is largely unexplored. This study describes the biosorption of Cd2+ by Sphingomonas sp. M1-B02, which was isolated from the moraine on the north slope of Mount Everest and has a good potential for biosorption. The difference in Cd2+ adsorption of the strain after UV irradiation stimulation indicated that the adsorption reached 68.90% in 24 h, but the adsorption after UV irradiation increased to 80.56%. The genome of strain M1-B02 contained antioxidant genes such as mutL, recA, recO, and heavy metal repair genes such as RS14805, apaG, chrA. Hydroxyl, nitro, and etceteras bonds on the bacterial surface were involved in Cd2+ adsorption through complexation reactions. The metabolites of the strains were significantly different after 24 h of Cd2+ stress, with pyocyanin, L-proline, hypoxanthine, etc., being downregulated and presumably involved in Cd2+ biosorption and upregulated after UV-C irradiation, which may explain the increase in Cd2+ adsorption capacity of the strain after UV-C irradiation, while the strain improved the metabolism of the antioxidant metabolite carnosine, indirectly increasing the adsorption capacity of the strains for Cd2+.
Keywords: Mount Everest; Sphingomonas; biosorption; heavy metal; metabolic analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Angeletti R., Binato G., Guidotti M., Morelli S., Pastorelli A.A., Sagratella E., Ciardullo S., Stacchini P. Cadmium Bioaccumulation in Mediterranean Spider Crab (Maya squinado): Human Consumption and Health Implications for Exposure in Italian Population. Chemosphere. 2014;100:83–88. doi: 10.1016/j.chemosphere.2013.12.056. - DOI - PubMed
-
- Ullah S., Hasan Z., Zuberi A. Heavy Metals in Three Commercially Valuable Cyprinids in the River Panjkora, District Lower Dir, Khyber Pakhtunkhwa, Pakistan. Toxicol. Environ. Chem. 2016;98:64–76. doi: 10.1080/02772248.2015.1100916. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

