Genomic Diversity and Virulence Factors of Clostridium perfringens Isolated from Healthy and Necrotic Enteritis-Affected Broiler Chicken Farms in Quebec Province
- PMID: 39770825
- PMCID: PMC11677781
- DOI: 10.3390/microorganisms12122624
Genomic Diversity and Virulence Factors of Clostridium perfringens Isolated from Healthy and Necrotic Enteritis-Affected Broiler Chicken Farms in Quebec Province
Abstract
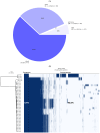
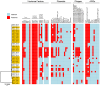
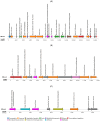
Avian necrotic enteritis due to the Gram-positive bacterium Clostridium perfringens has re-emerged following the ban on antibiotic growth promoters in many poultry producing countries. The limited number of previous studies has left important gaps in our understanding of the genetic diversity and virulence traits of the pathogen. To address these knowledge gaps, in this study, we sequenced the genomes of 41 Clostridium perfringens isolates recovered from commercial broiler chicken flocks in Quebec, Canada, including isolates from healthy birds and those affected by necrotic enteritis. We sought to understand the pangenome diversity and interrogated the genomes for key virulence factors involved in necrotic enteritis pathogenesis. On average, the genomes had a GC content of 28% and contained 3206 coding sequences. A variable presence of toxins, degradative hydrolytic enzymes, and collagen-binding proteins was also found. Through pangenome analysis, we revealed a total of 10,223 genes, 652 (6.4%) of which formed the core genome. Additionally, we identified 17 different plasmids, 12 antibiotic resistance genes, and nine prophage regions. Overall, our results demonstrated a relatively high genetic diversity among chicken Clostridium perfringens isolates collected from the same geographical location, offering new insights into potential virulence mechanisms and adaptation of the pathogen within poultry populations.
Keywords: Clostridium perfringens; Quebec; antibiotic resistance genes; broiler chickens; comparative genomics; necrotic enteritis; pangenome; prophage sequences; whole genome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Alizadeh M., Shojadoost B., Boodhoo N., Astill J., Taha-Abdelaziz K., Hodgins D.C., Kulkarni R.R., Sharif S. Necrotic enteritis in chickens: A review of pathogenesis, immune responses and prevention, focusing on probiotics and vaccination. Anim. Health Res. Rev. 2021;22:147–162. doi: 10.1017/S146625232100013X. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

