Comparative Genomics and Biosynthetic Cluster Analysis of Antifungal Secondary Metabolites of Three Strains of Streptomyces albidoflavus Isolated from Rhizospheric Soils
- PMID: 39770839
- PMCID: PMC11678301
- DOI: 10.3390/microorganisms12122637
Comparative Genomics and Biosynthetic Cluster Analysis of Antifungal Secondary Metabolites of Three Strains of Streptomyces albidoflavus Isolated from Rhizospheric Soils
Abstract
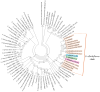
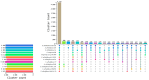
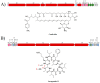
Streptomyces is a genus of Gram-positive bacteria with high GC content. It remains attractive for studying and discovering new antibiotics, antifungals, and chemotherapeutics. Streptomyces genomes can contain more than 30 cryptic and expressed biosynthetic gene clusters (BGC) encoding secondary metabolites. In this study, three Streptomyces strains isolated from jungle rhizospheric soil exhibited supernatants that can inhibit sensitive and fluconazole-resistant Candida spp. The genomes of the strains Streptomyces sp. A1, J25, J29 ori2 were sequenced, assembled de novo, and analyzed. The genome assemblies revealed that the size of the genomes was 6.9 Mb, with linear topology and 73.5% GC. A phylogenomic approach identified the strains with high similitudes between 98.5 and 98.7% with Streptomyces albidoflavus SM254 and R-53649 strains, respectively. Pangenomic analysis of eight genomes of S. albidoflavus strains deposited in the Genomes database recognized 4707 core protein orthogroups and 745 abundant accessory and exclusive protein orthogroups, suggesting an open pangenome in this species. The antiSMASH software detected candicidin and surugamide BGC-encoding polyene and octapeptide antifungal secondary metabolites in other S. albidoflavus. CORASON software was used to compare the synteny, and the abundance of genes harbored in the clusters was used. In conclusion, although the three strains belong to the same species, each possesses a distinct genome, as evidenced by the different phenotypes, including antifungal and extracellular enzymatic activities.
Keywords: BGC; CORASON; Candida; Streptomyces; antiSMASH; antifungal; candicidin; polyene; surugamide; synteny.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Streptomyces albidoflavus Q antifungal metabolites inhibit the ergosterol biosynthesis pathway and yeast growth in fluconazole-resistant Candida glabrata: phylogenomic and metabolomic analyses.Microbiol Spectr. 2023 Sep 27;11(5):e0127123. doi: 10.1128/spectrum.01271-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37754674 Free PMC article.
-
Candicidin Isomer Production Is Essential for Biocontrol of Cucumber Rhizoctonia Rot by Streptomyces albidoflavus W68.Appl Environ Microbiol. 2021 Apr 13;87(9):e03078-20. doi: 10.1128/AEM.03078-20. Print 2021 Apr 13. Appl Environ Microbiol. 2021. PMID: 33608297 Free PMC article.
-
Genome Mining Coupled with OSMAC-Based Cultivation Reveal Differential Production of Surugamide A by the Marine Sponge Isolate Streptomyces sp. SM17 When Compared to Its Terrestrial Relative S. albidoflavus J1074.Microorganisms. 2019 Sep 26;7(10):394. doi: 10.3390/microorganisms7100394. Microorganisms. 2019. PMID: 31561472 Free PMC article.
-
Comparative Genomics Reveals a Remarkable Biosynthetic Potential of the Streptomyces Phylogenetic Lineage Associated with Rugose-Ornamented Spores.mSystems. 2021 Aug 31;6(4):e0048921. doi: 10.1128/mSystems.00489-21. Epub 2021 Aug 24. mSystems. 2021. PMID: 34427515 Free PMC article.
-
Polyenic Antibiotics and Other Antifungal Compounds Produced by Hemolytic Streptomyces Species.Int J Mol Sci. 2022 Nov 30;23(23):15045. doi: 10.3390/ijms232315045. Int J Mol Sci. 2022. PMID: 36499372 Free PMC article. Review.
Cited by
-
Genomic insights into Streptomyces albidoflavus SM254: tracing the putative signs of anti-Pseudogymnoascus destructans properties.Braz J Microbiol. 2025 Sep;56(3):2121-2131. doi: 10.1007/s42770-025-01740-8. Epub 2025 Jul 22. Braz J Microbiol. 2025. PMID: 40694258 Free PMC article.
-
Streptomyces flavusporus sp. nov., a Novel Actinomycete Isolated from Naidong, Xizang (Tibet), China.Microorganisms. 2025 Apr 27;13(5):1001. doi: 10.3390/microorganisms13051001. Microorganisms. 2025. PMID: 40431174 Free PMC article.
References
-
- Youseif S.H., Abd El-Megeed F.H., Humm E.A., Maymon M., Mohamed A.H., Saleh S.A., Hirsch A.M. Comparative Analysis of the Cultured and Total Bacterial Community in the Wheat Rhizosphere Microbiome Using Culture-Dependent and Culture-Independent Approaches. Microbiol. Spectr. 2021;9:e0067821. doi: 10.1128/Spectrum.00678-21. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

