Association of Metabolic Diseases and Moderate Fat Intake with Myocardial Infarction Risk
- PMID: 39770895
- PMCID: PMC11679910
- DOI: 10.3390/nu16244273
Association of Metabolic Diseases and Moderate Fat Intake with Myocardial Infarction Risk
Abstract
Background: Myocardial infarction (MI) can range from mild to severe cardiovascular events and typically develops through complex interactions between genetic and lifestyle factors.
Objectives: We aimed to understand the genetic predisposition associated with MI through genetic correlation, colocalization analysis, and cells' gene expression values to develop more effective prevention and treatment strategies to reduce its burden.
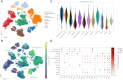
Methods: A polygenic risk score (PRS) was employed to estimate the genetic risk for MI and to analyze the dietary interactions with PRS that affect MI risk in adults over 45 years (n = 58,701). Genetic correlation (rg) between MI and metabolic syndrome-related traits was estimated with linkage disequilibrium score regression. Single-cell RNA sequencing (scRNA-seq) analysis was performed to investigate cellular heterogeneity in MI-associated genes.
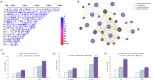
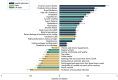
Results: Ten significant genetic variants associated with MI risk were related to cardiac, immune, and brain functions. A high PRS was associated with a threefold increase in MI risk (OR: 3.074, 95% CI: 2.354-4.014, p < 0.001). This increased the risk of MI plus obesity, hyperglycemia, dyslipidemia, and hypertension by about twofold after adjusting for MI-related covariates (p < 0.001). The PRS interacted with moderate fat intake (>15 energy percent), alcohol consumption (<30 g/day), and non-smoking, reducing MI risk in participants with a high PRS. MI was negatively correlated with the consumption of olive oil, sesame oil, and perilla oil used for cooking (rg = -0.364). MI risk was associated with storkhead box 1 (STOX1) and vacuolar protein sorting-associated protein 26A (VPS26A) in atrial and ventricular cardiomyocytes and fibroblasts.
Conclusions: This study identified novel genetic variants and gene expression patterns associated with MI risk, influenced by their interaction with fat and alcohol intake, and smoking status. Our findings provide insights for developing personalized prevention and treatment strategies targeting this complex clinical presentation of MI.
Keywords: fat; metabolic syndrome; myocardial infarction; polygenic risk score; precision medicine; single-cell RNA sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Height-Related Polygenic Variants Are Associated with Metabolic Syndrome Risk and Interact with Energy Intake and a Rice-Main Diet to Influence Height in KoGES.Nutrients. 2023 Apr 4;15(7):1764. doi: 10.3390/nu15071764. Nutrients. 2023. PMID: 37049604 Free PMC article.
-
Association between Polygenetic Risk Scores of Low Immunity and Interactions between These Scores and Moderate Fat Intake in a Large Cohort.Nutrients. 2021 Aug 19;13(8):2849. doi: 10.3390/nu13082849. Nutrients. 2021. PMID: 34445011 Free PMC article.
-
Gene-diet interactions in carbonated sugar-sweetened beverage consumption and metabolic syndrome risk: A machine learning analysis in a large hospital-based cohort.Clin Nutr ESPEN. 2024 Dec;64:358-369. doi: 10.1016/j.clnesp.2024.10.004. Epub 2024 Oct 11. Clin Nutr ESPEN. 2024. PMID: 39396701
-
Omega-6 fats for the primary and secondary prevention of cardiovascular disease.Cochrane Database Syst Rev. 2018 Nov 29;11(11):CD011094. doi: 10.1002/14651858.CD011094.pub4. Cochrane Database Syst Rev. 2018. PMID: 30488422 Free PMC article.
-
Reduction in saturated fat intake for cardiovascular disease.Cochrane Database Syst Rev. 2020 Aug 21;8(8):CD011737. doi: 10.1002/14651858.CD011737.pub3. Cochrane Database Syst Rev. 2020. PMID: 32827219 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

