Identification and Analysis of the Plasma Membrane H+-ATPase Gene Family in Cotton and Its Roles in Response to Salt Stress
- PMID: 39771208
- PMCID: PMC11728463
- DOI: 10.3390/plants13243510
Identification and Analysis of the Plasma Membrane H+-ATPase Gene Family in Cotton and Its Roles in Response to Salt Stress
Abstract
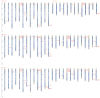
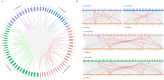
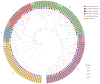
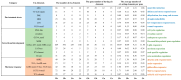
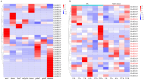
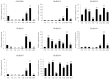
Plant plasma membrane (PM) H+-ATPase functions as a proton-motive force by exporting cellular protons to establish a transmembrane chemical gradient of H+ ions and an accompanying electrical gradient. These gradients are crucial for plant growth and development and for plant responses to abiotic and biotic stresses. In this study, a comprehensive identification of the PM H+-ATPase gene family was conducted across four cotton species. Specifically, 14 genes were identified in the diploid species Gossypium arboreum and Gossypium raimondii, whereas 39 and 43 genes were identified in the tetraploid species Gossypium hirsutum and Gossypium barbadense, respectively. The characteristics of this gene family were subsequently compared and analyzed using bioinformatics. Chromosomal localization and collinearity analyses elucidated the distribution characteristics of this gene family within the cotton genomes. Gene structure and phylogenetic analyses demonstrated the conservation of this family across cotton species, whereas the examination of cis-acting elements in gene promoters highlighted their involvement in environmental stress and hormone response categories. An expression profile analysis revealed eight genes whose expression was upregulated under salt stress conditions, and quantitative real-time PCR results suggested that the cotton PM H+-ATPase genes may play crucial roles in conferring resistance to salt stress. These findings establish a robust foundation for subsequent investigations into the functions of cotton PM H+-ATPase genes and may offer valuable insights for selecting genes for resistance breeding programs.
Keywords: PM H+-ATPase gene family; bioinformatics; cotton species; qRT-PCR; salt stress.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Genome-wide analyses of member identification, expression pattern, and protein-protein interaction of EPF/EPFL gene family in Gossypium.BMC Plant Biol. 2024 Jun 14;24(1):554. doi: 10.1186/s12870-024-05262-7. BMC Plant Biol. 2024. PMID: 38877405 Free PMC article.
-
Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species.BMC Genomics. 2017 Apr 12;18(1):292. doi: 10.1186/s12864-017-3682-x. BMC Genomics. 2017. PMID: 28403834 Free PMC article.
-
Genomic Comparison of the P-ATPase Gene Family in Four Cotton Species and Their Expression Patterns in Gossypium hirsutum.Molecules. 2018 May 5;23(5):1092. doi: 10.3390/molecules23051092. Molecules. 2018. PMID: 29734726 Free PMC article.
-
Genome wide identification, classification and functional characterization of heat shock transcription factors in cultivated and ancestral cottons (Gossypium spp.).Int J Biol Macromol. 2021 Jul 1;182:1507-1527. doi: 10.1016/j.ijbiomac.2021.05.016. Epub 2021 May 7. Int J Biol Macromol. 2021. PMID: 33965497
-
Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses.BMC Genomics. 2018 Aug 9;19(1):599. doi: 10.1186/s12864-018-4985-2. BMC Genomics. 2018. PMID: 30092779 Free PMC article.
Cited by
-
Genome-wide identification and analysis of the apple H+-ATPase gene family and its expression against iron deficiency stress.BMC Plant Biol. 2025 Apr 11;25(1):461. doi: 10.1186/s12870-025-06501-1. BMC Plant Biol. 2025. PMID: 40217534 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

